Batch loading data
Daniel Anderson
Week 4, Class 1
1 / 62
Example
- Create a function that simulates data (please copy the code and follow along)
set.seed(8675309)simulate <- function(n, mean = 0, sd = 1) { tibble(sample_id = seq_len(n), sample = rnorm(n, mean, sd))}simulate(10)## # A tibble: 10 x 2## sample_id sample## <int> <dbl>## 1 1 -0.9965824 ## 2 2 0.7218241 ## 3 3 -0.6172088 ## 4 4 2.029392 ## 5 5 1.065416 ## 6 6 0.9872197 ## 7 7 0.02745393## 8 8 0.6728723 ## 9 9 0.5720665 ## 10 10 0.90367777 / 62
Two more quick examples
simulate(3, 100, 10)## # A tibble: 3 x 2## sample_id sample## <int> <dbl>## 1 1 84.50448## 2 2 110.2264 ## 3 3 101.5008simulate(5, -10, 1.5)## # A tibble: 5 x 2## sample_id sample## <int> <dbl>## 1 1 -10.98995 ## 2 2 -11.49188 ## 3 3 -7.041312## 4 4 -10.66270 ## 5 5 -11.350968 / 62
library(tidyverse)sims <- map(seq(10, 150, 5), simulate, 100, 10)sims[1]## [[1]]## # A tibble: 10 x 2## sample_id sample## <int> <dbl>## 1 1 103.7618 ## 2 2 111.5353 ## 3 3 115.7490 ## 4 4 105.8853 ## 5 5 93.84955## 6 6 97.71089## 7 7 100.6392 ## 8 8 96.86526## 9 9 97.51501## 10 10 98.46205sims[2]## [[1]]## # A tibble: 15 x 2## sample_id sample## <int> <dbl>## 1 1 93.64743## 2 2 99.96206## 3 3 100.4562 ## 4 4 106.8407 ## 5 5 97.47957## 6 6 98.48961## 7 7 91.25069## 8 8 80.23099## 9 9 102.3766 ## 10 10 100.3609 ## 11 11 101.3490 ## 12 12 101.1758 ## 13 13 91.74411## 14 14 78.64764## 15 15 102.142110 / 62
Swap for map_dfr
Try it - what happens?
01:00
sims_df <- map_dfr(seq(10, 150, 5), simulate, 100, 10)sims_df## # A tibble: 2,320 x 2## sample_id sample## <int> <dbl>## 1 1 85.64361## 2 2 103.6789 ## 3 3 94.71782## 4 4 103.1350 ## 5 5 99.78701## 6 6 105.3462 ## 7 7 100.0653 ## 8 8 94.28314## 9 9 108.8872 ## 10 10 106.0850 ## # … with 2,310 more rows11 / 62
Notice a problem here
sims_df[1:15, ]## # A tibble: 15 x 2## sample_id sample## <int> <dbl>## 1 1 85.64361## 2 2 103.6789 ## 3 3 94.71782## 4 4 103.1350 ## 5 5 99.78701## 6 6 105.3462 ## 7 7 100.0653 ## 8 8 94.28314## 9 9 108.8872 ## 10 10 106.0850 ## 11 1 89.49968## 12 2 86.99898## 13 3 85.38054## 14 4 99.10690## 15 5 105.008812 / 62
.id argument
sims_df2 <- map_dfr(seq(10, 150, 5), simulate, 100, 10, .id = "iteration")sims_df2[1:14, ]## # A tibble: 14 x 3## iteration sample_id sample## <chr> <int> <dbl>## 1 1 1 112.1250 ## 2 1 2 88.07056## 3 1 3 108.3908 ## 4 1 4 100.8193 ## 5 1 5 102.1545 ## 6 1 6 113.5398 ## 7 1 7 101.4171 ## 8 1 8 99.33668## 9 1 9 100.2855 ## 10 1 10 90.22043## 11 2 1 91.08882## 12 2 2 107.3664 ## 13 2 3 101.1745 ## 14 2 4 96.8205313 / 62
.id: Either a string or NULL. If a string, the output will contain a variable with that name, storing either the name (if .x is named) or the index (if .x is unnamed) of the input. If NULL, the default, no variable will be created.
14 / 62
setNames
sample_size <- seq(10, 150, 5)sample_size## [1] 10 15 20 25 30 35 40 45 50 55 60 65 70 75 80 85 90 95 100 105 110 115## [23] 120 125 130 135 140 145 150sample_size <- setNames(sample_size, english::english(seq(10, 150, 5))) sample_size[1:15]## ten fifteen twenty twenty-five thirty thirty-five forty ## 10 15 20 25 30 35 40 ## forty-five fifty fifty-five sixty sixty-five seventy seventy-five ## 45 50 55 60 65 70 75 ## eighty ## 8015 / 62
Try again
sims_df3 <- map_dfr(sample_size, simulate, 100, 10, .id = "n")sims_df3[1:14, ]## # A tibble: 14 x 3## n sample_id sample## <chr> <int> <dbl>## 1 ten 1 98.94914## 2 ten 2 101.6824 ## 3 ten 3 88.16447## 4 ten 4 90.13604## 5 ten 5 85.53591## 6 ten 6 90.69977## 7 ten 7 105.8858 ## 8 ten 8 89.12978## 9 ten 9 114.4982 ## 10 ten 10 111.6440 ## 11 fifteen 1 103.2732 ## 12 fifteen 2 106.8949 ## 13 fifteen 3 88.83591## 14 fifteen 4 105.540216 / 62
Another quick example
broom::tidy
- The {broom} package helps us extract model output in a tidy format
lm(tvhours ~ age, gss_cat) %>% broom::tidy()## # A tibble: 2 x 5## term estimate std.error statistic p.value## <chr> <dbl> <dbl> <dbl> <dbl>## 1 (Intercept) 1.992391 0.06980966 28.54033 4.672161e-173## 2 age 0.02095679 0.001387361 15.10551 4.689310e- 5117 / 62
Fit separate models by year
Again - probs not best statistically
split(gss_cat, gss_cat$year) %>% map_dfr(~lm(tvhours ~ age, .x) %>% broom::tidy())## # A tibble: 16 x 5## term estimate std.error statistic p.value## <chr> <dbl> <dbl> <dbl> <dbl>## 1 (Intercept) 2.080163 0.1709061 12.17138 7.995632e-33## 2 age 0.01948584 0.003485199 5.591027 2.599011e- 8## 3 (Intercept) 2.078999 0.2176829 9.550583 1.191266e-20## 4 age 0.01963575 0.004400292 4.462375 9.137366e- 6## 5 (Intercept) 1.767990 0.2464509 7.173804 1.531756e-12## 6 age 0.02386070 0.005031548 4.742218 2.459650e- 6## 7 (Intercept) 2.096054 0.1496431 14.00702 1.419772e-42## 8 age 0.01781388 0.002977289 5.983256 2.589482e- 9## 9 (Intercept) 1.855278 0.2156381 8.603668 2.167351e-17## 10 age 0.02390720 0.004314567 5.541043 3.628675e- 8## 11 (Intercept) 2.068914 0.2096397 9.868903 2.896085e-22## 12 age 0.01989505 0.004086638 4.868317 1.251234e- 6## 13 (Intercept) 1.878070 0.2258400 8.315932 2.280108e-16## 14 age 0.02547794 0.004449295 5.726287 1.274840e- 8## 15 (Intercept) 1.980095 0.1877544 10.54620 3.238043e-25## 16 age 0.02049066 0.003611900 5.673098 1.650822e- 818 / 62
.id
In cases like the preceding, .id becomes invaluable
split(gss_cat, gss_cat$year) %>% map_dfr(~lm(tvhours ~ age, .x) %>% broom::tidy(), .id = "year")## # A tibble: 16 x 6## year term estimate std.error statistic p.value## <chr> <chr> <dbl> <dbl> <dbl> <dbl>## 1 2000 (Intercept) 2.080163 0.1709061 12.17138 7.995632e-33## 2 2000 age 0.01948584 0.003485199 5.591027 2.599011e- 8## 3 2002 (Intercept) 2.078999 0.2176829 9.550583 1.191266e-20## 4 2002 age 0.01963575 0.004400292 4.462375 9.137366e- 6## 5 2004 (Intercept) 1.767990 0.2464509 7.173804 1.531756e-12## 6 2004 age 0.02386070 0.005031548 4.742218 2.459650e- 6## 7 2006 (Intercept) 2.096054 0.1496431 14.00702 1.419772e-42## 8 2006 age 0.01781388 0.002977289 5.983256 2.589482e- 9## 9 2008 (Intercept) 1.855278 0.2156381 8.603668 2.167351e-17## 10 2008 age 0.02390720 0.004314567 5.541043 3.628675e- 8## 11 2010 (Intercept) 2.068914 0.2096397 9.868903 2.896085e-22## 12 2010 age 0.01989505 0.004086638 4.868317 1.251234e- 6## 13 2012 (Intercept) 1.878070 0.2258400 8.315932 2.280108e-16## 14 2012 age 0.02547794 0.004449295 5.726287 1.274840e- 8## 15 2014 (Intercept) 1.980095 0.1877544 10.54620 3.238043e-25## 16 2014 age 0.02049066 0.003611900 5.673098 1.650822e- 819 / 62
{fs}
- note - there are base equivalents.
{fs}is just a a bit better across platforms and has better defaults.
Could we apply map_dfr here?
# install.packages("fs")library(fs)dir_ls(here::here("data"))## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_simdir_ls(here::here("data", "pfiles_sim"))## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g11ELApfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g11Mathpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g11Rdgpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g11Sciencepfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g11Wripfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g3ELApfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g3Mathpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g3Rdgpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g3Wripfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g4ELApfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g4Mathpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g4Rdgpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g4Wripfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g5ELApfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g5Mathpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g5Rdgpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g5Sciencepfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g5Wripfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g6ELApfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g6Mathpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g6Rdgpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g6Wripfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g7ELApfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g7Mathpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g7Rdgpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g7Wripfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g8ELApfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g8Mathpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g8Rdgpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g8Sciencepfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g8Wripfiles18_sim.csv21 / 62
Limit files
- We really only want the
.csv- That happens to be the only thing that's in there but that's regularly not the case
dir_ls(here::here("data", "pfiles_sim"), glob = "*.csv")## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g11ELApfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g11Mathpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g11Rdgpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g11Sciencepfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g11Wripfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g3ELApfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g3Mathpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g3Rdgpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g3Wripfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g4ELApfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g4Mathpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g4Rdgpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g4Wripfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g5ELApfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g5Mathpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g5Rdgpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g5Sciencepfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g5Wripfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g6ELApfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g6Mathpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g6Rdgpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g6Wripfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g7ELApfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g7Mathpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g7Rdgpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g7Wripfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g8ELApfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g8Mathpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g8Rdgpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g8Sciencepfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g8Wripfiles18_sim.csv22 / 62
Batch load
- Loop through the directories and
importorread_csv
files <- dir_ls( here::here("data", "pfiles_sim"), glob = "*.csv")batch <- map_dfr(files, read_csv)batch## # A tibble: 15,945 x 22## Entry Theta Status Count RawScore SE Infit Infit_Z Outfit Outfit_Z Displacement## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 123 1.2687 1 36 23 0.3713 0.93 -0.34 0.82 -0.62 0.0018## 2 88 1.5541 1 36 25 0.3852 0.95 -0.37 0.81 -0.56 0.0019## 3 105 3.2773 1 36 33 0.6187 0.9 -0.04 1.63 1.03 0.0022## 4 153 4.4752 1 36 35 1.0234 0.93 0.23 0.35 -0.16 0.0023## 5 437 2.6655 1 36 31 0.5008 0.92 -0.18 0.88 -0.12 0.0021## 6 307 5.7137 0 36 36 1.8371 1 0 1 0 0.0024## 7 305 3.7326 1 36 34 0.7408 1.06 0.31 0.86 0.17 0.0022## 8 42 0.609 1 36 18 0.36 1.55 2.56 1.74 3.31 0.0017## 9 59 -2.623 1 36 3 1.0344 0.85 0.06 0.17 -0.37 0.0009## 10 304 5.7137 0 36 36 1.8371 1 0 1 0 0.0024## # … with 15,935 more rows, and 11 more variables: PointMeasureCorr <dbl>, Weight <dbl>,## # ObservMatch <dbl>, ExpectMatch <dbl>, PointMeasureExpected <dbl>, RMSR <dbl>,## # WMLE <dbl>, testeventid <dbl>, ssid <dbl>, asmtprmrydsbltycd <dbl>,## # asmtscndrydsbltycd <dbl>23 / 62
Add id
batch2 <- map_dfr(files, read_csv, .id = "file")batch2## # A tibble: 15,945 x 23## # … with 15,935 more rows, and 23 more variables: file <chr>, Entry <dbl>, Theta <dbl>,## # Status <dbl>, Count <dbl>, RawScore <dbl>, SE <dbl>, Infit <dbl>, Infit_Z <dbl>,## # Outfit <dbl>, Outfit_Z <dbl>, Displacement <dbl>, PointMeasureCorr <dbl>, Weight <dbl>,## # ObservMatch <dbl>, ExpectMatch <dbl>, PointMeasureExpected <dbl>, RMSR <dbl>,## # WMLE <dbl>, testeventid <dbl>, ssid <dbl>, asmtprmrydsbltycd <dbl>,## # asmtscndrydsbltycd <dbl>Note - the file column contains the full path, which is so long it makes no rows print
25 / 62
Step 1
- Remove the
here::herepath from string
batch2 <- batch2 %>% mutate( file = str_replace_all( file, here::here("data", "pfiles_sim"), "" ) )batch2 %>% count(file)## # A tibble: 31 x 2## file n## <chr> <int>## 1 /g11ELApfiles18_sim.csv 453## 2 /g11Mathpfiles18_sim.csv 460## 3 /g11Rdgpfiles18_sim.csv 453## 4 /g11Sciencepfiles18_sim.csv 438## 5 /g11Wripfiles18_sim.csv 453## 6 /g3ELApfiles18_sim.csv 540## 7 /g3Mathpfiles18_sim.csv 536## 8 /g3Rdgpfiles18_sim.csv 540## 9 /g3Wripfiles18_sim.csv 540## 10 /g4ELApfiles18_sim.csv 585## # … with 21 more rows27 / 62
Pull out pieces you need
Regular expressions are most powerful here
- We haven't talked about them much
Try RegExplain
28 / 62
Pull grade
- Note - I'm not expecting you to just suddenly be able to do this. This is more for illustration. There's also other ways you could extract the same info
batch2 %>% mutate( grade = str_replace_all( file, "/g(\\d?\\d).+", "\\1" ) ) %>% select(file, grade)## # A tibble: 15,945 x 2## file grade## <chr> <chr>## 1 /g11ELApfiles18_sim.csv 11 ## 2 /g11ELApfiles18_sim.csv 11 ## 3 /g11ELApfiles18_sim.csv 11 ## 4 /g11ELApfiles18_sim.csv 11 ## 5 /g11ELApfiles18_sim.csv 11 ## 6 /g11ELApfiles18_sim.csv 11 ## 7 /g11ELApfiles18_sim.csv 11 ## 8 /g11ELApfiles18_sim.csv 11 ## 9 /g11ELApfiles18_sim.csv 11 ## 10 /g11ELApfiles18_sim.csv 11 ## # … with 15,935 more rows29 / 62
parse_number
- In this case
parse_numberalso works - but note that it would not work to extract the year
batch2 %>%mutate(grade = parse_number(file)) %>% select(file, grade)## # A tibble: 15,945 x 2## file grade## <chr> <dbl>## 1 /g11ELApfiles18_sim.csv 11## 2 /g11ELApfiles18_sim.csv 11## 3 /g11ELApfiles18_sim.csv 11## 4 /g11ELApfiles18_sim.csv 11## 5 /g11ELApfiles18_sim.csv 11## 6 /g11ELApfiles18_sim.csv 11## 7 /g11ELApfiles18_sim.csv 11## 8 /g11ELApfiles18_sim.csv 11## 9 /g11ELApfiles18_sim.csv 11## 10 /g11ELApfiles18_sim.csv 11## # … with 15,935 more rows30 / 62
Extract year
- In this case
parse_numberalso works - but note that it would not work to extract the year
batch2 %>% mutate( grade = str_replace_all( file, "/g(\\d?\\d).+", "\\1" ), year = str_replace_all( file, ".+files(\\d\\d)_sim.+", "\\1" ) ) %>% select(file, grade, year)## # A tibble: 15,945 x 3## file grade year ## <chr> <chr> <chr>## 1 /g11ELApfiles18_sim.csv 11 18 ## 2 /g11ELApfiles18_sim.csv 11 18 ## 3 /g11ELApfiles18_sim.csv 11 18 ## 4 /g11ELApfiles18_sim.csv 11 18 ## 5 /g11ELApfiles18_sim.csv 11 18 ## 6 /g11ELApfiles18_sim.csv 11 18 ## 7 /g11ELApfiles18_sim.csv 11 18 ## 8 /g11ELApfiles18_sim.csv 11 18 ## 9 /g11ELApfiles18_sim.csv 11 18 ## 10 /g11ELApfiles18_sim.csv 11 18 ## # … with 15,935 more rows31 / 62
Extract Content Area
batch2 %>% mutate(grade = str_replace_all(file, "/g(\\d?\\d).+", "\\1"), year = str_replace_all(file, ".+files(\\d\\d)_sim.+", "\\1"), content = str_replace_all(file, "/g\\d?\\d(.+)pfiles.+", "\\1")) %>% select(file, grade, year, content)## # A tibble: 15,945 x 4## file grade year content## <chr> <chr> <chr> <chr> ## 1 /g11ELApfiles18_sim.csv 11 18 ELA ## 2 /g11ELApfiles18_sim.csv 11 18 ELA ## 3 /g11ELApfiles18_sim.csv 11 18 ELA ## 4 /g11ELApfiles18_sim.csv 11 18 ELA ## 5 /g11ELApfiles18_sim.csv 11 18 ELA ## 6 /g11ELApfiles18_sim.csv 11 18 ELA ## 7 /g11ELApfiles18_sim.csv 11 18 ELA ## 8 /g11ELApfiles18_sim.csv 11 18 ELA ## 9 /g11ELApfiles18_sim.csv 11 18 ELA ## 10 /g11ELApfiles18_sim.csv 11 18 ELA ## # … with 15,935 more rows32 / 62
Double checks: grade
batch2 %>% mutate(grade = str_replace_all(file, "/g(\\d?\\d).+", "\\1"), year = str_replace_all(file, ".+files(\\d\\d)_sim.+", "\\1"), content = str_replace_all(file, "/g\\d?\\d(.+)pfiles.+", "\\1")) %>% select(file, grade, year, content) %>% count(grade)## # A tibble: 7 x 2## grade n## <chr> <int>## 1 11 2257## 2 3 2156## 3 4 2341## 4 5 2632## 5 6 2216## 6 7 1962## 7 8 238133 / 62
Double checks: year
batch2 %>% mutate(grade = str_replace_all(file, "/g(\\d?\\d).+", "\\1"), year = str_replace_all(file, ".+files(\\d\\d)_sim.+", "\\1"), content = str_replace_all(file, "/g\\d?\\d(.+)pfiles.+", "\\1")) %>% select(file, grade, year, content) %>% count(year)## # A tibble: 1 x 2## year n## <chr> <int>## 1 18 1594534 / 62
Double checks: content
batch2 %>% mutate(grade = str_replace_all(file, "/g(\\d?\\d).+", "\\1"), year = str_replace_all(file, ".+files(\\d\\d)_sim.+", "\\1"), content = str_replace_all(file, "/g\\d?\\d(.+)pfiles.+", "\\1")) %>% select(file, grade, year, content) %>% count(content)## # A tibble: 5 x 2## content n## <chr> <int>## 1 ELA 3627## 2 Math 3629## 3 Rdg 3627## 4 Science 1435## 5 Wri 362735 / 62
Finalize
d <- batch2 %>% mutate(grade = str_replace_all(file, "/g(\\d?\\d).+", "\\1"), grade = as.integer(grade), year = str_replace_all(file, ".+files(\\d\\d)_sim.+", "\\1"), year = as.integer(grade), content = str_replace_all(file, "/g\\d?\\d(.+)pfiles.+", "\\1")) %>% select(-file) %>% select(ssid, grade, year, content, testeventid, asmtprmrydsbltycd, asmtscndrydsbltycd, Entry:WMLE)36 / 62
Final product
In this case, we basically have a tidy data frame already!
We've reduced our problem from 31 files to a single file
d## # A tibble: 15,945 x 25## ssid grade year content testeventid asmtprmrydsbltycd asmtscndrydsbltycd Entry## <dbl> <int> <int> <chr> <dbl> <dbl> <dbl> <dbl>## 1 9466908 11 11 ELA 148933 0 0 123## 2 7683685 11 11 ELA 147875 10 0 88## 3 9025693 11 11 ELA 143699 40 20 105## 4 10099824 11 11 ELA 143962 82 0 153## 5 18886078 11 11 ELA 150680 10 0 437## 6 10606750 11 11 ELA 144583 80 80 307## 7 10541306 11 11 ELA 145204 50 0 305## 8 7632967 11 11 ELA 148926 10 0 42## 9 7661118 11 11 ELA 148893 50 0 59## 10 10547177 11 11 ELA 144583 82 0 304## # … with 15,935 more rows, and 17 more variables: Theta <dbl>, Status <dbl>, Count <dbl>,## # RawScore <dbl>, SE <dbl>, Infit <dbl>, Infit_Z <dbl>, Outfit <dbl>, Outfit_Z <dbl>,## # Displacement <dbl>, PointMeasureCorr <dbl>, Weight <dbl>, ObservMatch <dbl>,## # ExpectMatch <dbl>, PointMeasureExpected <dbl>, RMSR <dbl>, WMLE <dbl>37 / 62
Summary stats
d %>% group_by(grade, content, asmtprmrydsbltycd) %>% summarize(mean = mean(Theta)) %>% pivot_wider(names_from = content, values_from = mean)## # A tibble: 77 x 7## # Groups: grade [7]## grade asmtprmrydsbltycd ELA Math Rdg Wri Science## <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 3 0 -0.07361 -1.21055 1.010455 1.612308 NA## 2 3 10 0.3700416 -0.8182091 0.5184354 0.3206475 NA## 3 3 20 -0.06335 -1.2514 1.52 -0.5775 NA## 4 3 40 -1.877683 -3.56365 -1.761667 -0.7514286 NA## 5 3 50 0.9462857 -0.09186957 0.9791176 1.191481 NA## 6 3 60 0.840775 1.040375 2.181111 1.067 NA## 7 3 70 -1.104049 -1.517955 -0.8454839 -1.005625 NA## 8 3 74 0.996 0.0208375 0.6 1.2925 NA## 9 3 80 -0.144304 -0.5325596 0.6791667 0.2686301 NA## 10 3 82 0.3708244 -1.080988 0.5676650 0.3440741 NA## # … with 67 more rows39 / 62
Backing up a bit
- What if we wanted only math files?
dir_ls(here::here("data", "pfiles_sim"), regexp = "Math")## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g11Mathpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g3Mathpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g4Mathpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g5Mathpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g6Mathpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g7Mathpfiles18_sim.csv## /Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g8Mathpfiles18_sim.csv40 / 62
The rest is the same
g5 <- map_dfr(g5_paths, read_csv, .id = "file") %>% mutate( file = str_replace_all( file, here::here("data", "pfiles_sim"), "" ) )g5## # A tibble: 2,632 x 23## file Entry Theta Status Count RawScore SE Infit Infit_Z Outfit## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 /g5ELApfiles18_sim.csv 375 3.154 1 36 32 0.551 1.22 0.91 2.1 ## 2 /g5ELApfiles18_sim.csv 305 0.3662 1 36 16 0.3894 0.94 -0.31 0.98## 3 /g5ELApfiles18_sim.csv 163 -4.9547 -1 36 0 1.8495 1 0 1 ## 4 /g5ELApfiles18_sim.csv 524 -4.9547 -1 36 0 1.8495 1 0 1 ## 5 /g5ELApfiles18_sim.csv 81 3.154 1 36 32 0.551 0.96 0 0.58## 6 /g5ELApfiles18_sim.csv 325 1.7156 1 36 25 0.3997 1.14 0.92 1.23## 7 /g5ELApfiles18_sim.csv 163 1.8786 1 36 26 0.4078 0.82 -1.08 0.57## 8 /g5ELApfiles18_sim.csv 116 5.9323 0 36 36 1.8373 1 0 1 ## 9 /g5ELApfiles18_sim.csv 273 1.4052 1 36 23 0.3891 1.08 0.44 0.94## 10 /g5ELApfiles18_sim.csv 202 1.8786 1 36 26 0.4078 0.79 -0.84 0.76## # … with 2,622 more rows, and 13 more variables: Outfit_Z <dbl>, Displacement <dbl>,## # PointMeasureCorr <dbl>, Weight <dbl>, ObservMatch <dbl>, ExpectMatch <dbl>,## # PointMeasureExpected <dbl>, RMSR <dbl>, WMLE <dbl>, testeventid <dbl>, ssid <dbl>,## # asmtprmrydsbltycd <dbl>, asmtscndrydsbltycd <dbl>42 / 62
Base equivalents
list.files(here::here("data", "pfiles_sim"))## [1] "g11ELApfiles18_sim.csv" "g11Mathpfiles18_sim.csv" "g11Rdgpfiles18_sim.csv" ## [4] "g11Sciencepfiles18_sim.csv" "g11Wripfiles18_sim.csv" "g3ELApfiles18_sim.csv" ## [7] "g3Mathpfiles18_sim.csv" "g3Rdgpfiles18_sim.csv" "g3Wripfiles18_sim.csv" ## [10] "g4ELApfiles18_sim.csv" "g4Mathpfiles18_sim.csv" "g4Rdgpfiles18_sim.csv" ## [13] "g4Wripfiles18_sim.csv" "g5ELApfiles18_sim.csv" "g5Mathpfiles18_sim.csv" ## [16] "g5Rdgpfiles18_sim.csv" "g5Sciencepfiles18_sim.csv" "g5Wripfiles18_sim.csv" ## [19] "g6ELApfiles18_sim.csv" "g6Mathpfiles18_sim.csv" "g6Rdgpfiles18_sim.csv" ## [22] "g6Wripfiles18_sim.csv" "g7ELApfiles18_sim.csv" "g7Mathpfiles18_sim.csv" ## [25] "g7Rdgpfiles18_sim.csv" "g7Wripfiles18_sim.csv" "g8ELApfiles18_sim.csv" ## [28] "g8Mathpfiles18_sim.csv" "g8Rdgpfiles18_sim.csv" "g8Sciencepfiles18_sim.csv" ## [31] "g8Wripfiles18_sim.csv"43 / 62
Full path
list.files(here::here("data", "pfiles_sim"), full.names = TRUE)## [1] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g11ELApfiles18_sim.csv" ## [2] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g11Mathpfiles18_sim.csv" ## [3] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g11Rdgpfiles18_sim.csv" ## [4] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g11Sciencepfiles18_sim.csv"## [5] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g11Wripfiles18_sim.csv" ## [6] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g3ELApfiles18_sim.csv" ## [7] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g3Mathpfiles18_sim.csv" ## [8] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g3Rdgpfiles18_sim.csv" ## [9] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g3Wripfiles18_sim.csv" ## [10] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g4ELApfiles18_sim.csv" ## [11] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g4Mathpfiles18_sim.csv" ## [12] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g4Rdgpfiles18_sim.csv" ## [13] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g4Wripfiles18_sim.csv" ## [14] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g5ELApfiles18_sim.csv" ## [15] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g5Mathpfiles18_sim.csv" ## [16] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g5Rdgpfiles18_sim.csv" ## [17] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g5Sciencepfiles18_sim.csv" ## [18] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g5Wripfiles18_sim.csv" ## [19] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g6ELApfiles18_sim.csv" ## [20] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g6Mathpfiles18_sim.csv" ## [21] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g6Rdgpfiles18_sim.csv" ## [22] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g6Wripfiles18_sim.csv" ## [23] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g7ELApfiles18_sim.csv" ## [24] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g7Mathpfiles18_sim.csv" ## [25] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g7Rdgpfiles18_sim.csv" ## [26] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g7Wripfiles18_sim.csv" ## [27] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g8ELApfiles18_sim.csv" ## [28] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g8Mathpfiles18_sim.csv" ## [29] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g8Rdgpfiles18_sim.csv" ## [30] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g8Sciencepfiles18_sim.csv" ## [31] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g8Wripfiles18_sim.csv"44 / 62
Only csvs
list.files(here::here("data", "pfiles_sim"), full.names = TRUE, pattern = "*.csv")## [1] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g11ELApfiles18_sim.csv" ## [2] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g11Mathpfiles18_sim.csv" ## [3] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g11Rdgpfiles18_sim.csv" ## [4] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g11Sciencepfiles18_sim.csv"## [5] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g11Wripfiles18_sim.csv" ## [6] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g3ELApfiles18_sim.csv" ## [7] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g3Mathpfiles18_sim.csv" ## [8] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g3Rdgpfiles18_sim.csv" ## [9] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g3Wripfiles18_sim.csv" ## [10] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g4ELApfiles18_sim.csv" ## [11] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g4Mathpfiles18_sim.csv" ## [12] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g4Rdgpfiles18_sim.csv" ## [13] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g4Wripfiles18_sim.csv" ## [14] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g5ELApfiles18_sim.csv" ## [15] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g5Mathpfiles18_sim.csv" ## [16] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g5Rdgpfiles18_sim.csv" ## [17] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g5Sciencepfiles18_sim.csv" ## [18] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g5Wripfiles18_sim.csv" ## [19] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g6ELApfiles18_sim.csv" ## [20] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g6Mathpfiles18_sim.csv" ## [21] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g6Rdgpfiles18_sim.csv" ## [22] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g6Wripfiles18_sim.csv" ## [23] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g7ELApfiles18_sim.csv" ## [24] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g7Mathpfiles18_sim.csv" ## [25] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g7Rdgpfiles18_sim.csv" ## [26] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g7Wripfiles18_sim.csv" ## [27] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g8ELApfiles18_sim.csv" ## [28] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g8Mathpfiles18_sim.csv" ## [29] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g8Rdgpfiles18_sim.csv" ## [30] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g8Sciencepfiles18_sim.csv" ## [31] "/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/data/pfiles_sim/g8Wripfiles18_sim.csv"45 / 62
Why not use base?
- We could, but
{fs}plays a little nicer with{purrr}
files <- list.files( here::here("data", "pfiles_sim"), pattern = "*.csv")batch3 <- map_dfr(files, read_csv, .id = "file")## Error: 'g11ELApfiles18_sim.csv' does not exist in current working directory ('/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/static/slides').46 / 62
Why not use base?
- We could, but
{fs}plays a little nicer with{purrr}
files <- list.files( here::here("data", "pfiles_sim"), pattern = "*.csv")batch3 <- map_dfr(files, read_csv, .id = "file")## Error: 'g11ELApfiles18_sim.csv' does not exist in current working directory ('/Users/daniel/Teaching/data_sci_specialization/2020-21/c3-fp-2021/static/slides').- Need to return full names
files## [1] "g11ELApfiles18_sim.csv" "g11Mathpfiles18_sim.csv" "g11Rdgpfiles18_sim.csv" ## [4] "g11Sciencepfiles18_sim.csv" "g11Wripfiles18_sim.csv" "g3ELApfiles18_sim.csv" ## [7] "g3Mathpfiles18_sim.csv" "g3Rdgpfiles18_sim.csv" "g3Wripfiles18_sim.csv" ## [10] "g4ELApfiles18_sim.csv" "g4Mathpfiles18_sim.csv" "g4Rdgpfiles18_sim.csv" ## [13] "g4Wripfiles18_sim.csv" "g5ELApfiles18_sim.csv" "g5Mathpfiles18_sim.csv" ## [16] "g5Rdgpfiles18_sim.csv" "g5Sciencepfiles18_sim.csv" "g5Wripfiles18_sim.csv" ## [19] "g6ELApfiles18_sim.csv" "g6Mathpfiles18_sim.csv" "g6Rdgpfiles18_sim.csv" ## [22] "g6Wripfiles18_sim.csv" "g7ELApfiles18_sim.csv" "g7Mathpfiles18_sim.csv" ## [25] "g7Rdgpfiles18_sim.csv" "g7Wripfiles18_sim.csv" "g8ELApfiles18_sim.csv" ## [28] "g8Mathpfiles18_sim.csv" "g8Rdgpfiles18_sim.csv" "g8Sciencepfiles18_sim.csv" ## [31] "g8Wripfiles18_sim.csv"46 / 62
Try again
files <- list.files(here::here("data", "pfiles_sim"), pattern = "*.csv", full.names = TRUE)batch3 <- map_dfr(files, read_csv, .id = "file")batch3## # A tibble: 15,945 x 23## file Entry Theta Status Count RawScore SE Infit Infit_Z Outfit Outfit_Z## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 1 123 1.2687 1 36 23 0.3713 0.93 -0.34 0.82 -0.62## 2 1 88 1.5541 1 36 25 0.3852 0.95 -0.37 0.81 -0.56## 3 1 105 3.2773 1 36 33 0.6187 0.9 -0.04 1.63 1.03## 4 1 153 4.4752 1 36 35 1.0234 0.93 0.23 0.35 -0.16## 5 1 437 2.6655 1 36 31 0.5008 0.92 -0.18 0.88 -0.12## 6 1 307 5.7137 0 36 36 1.8371 1 0 1 0 ## 7 1 305 3.7326 1 36 34 0.7408 1.06 0.31 0.86 0.17## 8 1 42 0.609 1 36 18 0.36 1.55 2.56 1.74 3.31## 9 1 59 -2.623 1 36 3 1.0344 0.85 0.06 0.17 -0.37## 10 1 304 5.7137 0 36 36 1.8371 1 0 1 0 ## # … with 15,935 more rows, and 12 more variables: Displacement <dbl>,## # PointMeasureCorr <dbl>, Weight <dbl>, ObservMatch <dbl>, ExpectMatch <dbl>,## # PointMeasureExpected <dbl>, RMSR <dbl>, WMLE <dbl>, testeventid <dbl>, ssid <dbl>,## # asmtprmrydsbltycd <dbl>, asmtscndrydsbltycd <dbl>47 / 62
Base method that works
files <- list.files(here::here("data", "pfiles_sim"), pattern = "*.csv", full.names = TRUE) files <- setNames(files, files)batch4 <- map_dfr(files, read_csv, .id = "file")batch4## # A tibble: 15,945 x 23## # … with 15,935 more rows, and 23 more variables: file <chr>, Entry <dbl>, Theta <dbl>,## # Status <dbl>, Count <dbl>, RawScore <dbl>, SE <dbl>, Infit <dbl>, Infit_Z <dbl>,## # Outfit <dbl>, Outfit_Z <dbl>, Displacement <dbl>, PointMeasureCorr <dbl>, Weight <dbl>,## # ObservMatch <dbl>, ExpectMatch <dbl>, PointMeasureExpected <dbl>, RMSR <dbl>,## # WMLE <dbl>, testeventid <dbl>, ssid <dbl>, asmtprmrydsbltycd <dbl>,## # asmtscndrydsbltycd <dbl>49 / 62
Split the data
The base method we've been using...
splt_content <- split(d, d$content)str(splt_content)## List of 5## $ ELA : tibble[,25] [3,627 × 25] (S3: tbl_df/tbl/data.frame)## ..$ ssid : num [1:3627] 9466908 7683685 9025693 10099824 18886078 ...## ..$ grade : int [1:3627] 11 11 11 11 11 11 11 11 11 11 ...## ..$ year : int [1:3627] 11 11 11 11 11 11 11 11 11 11 ...## ..$ content : chr [1:3627] "ELA" "ELA" "ELA" "ELA" ...## ..$ testeventid : num [1:3627] 148933 147875 143699 143962 150680 ...## ..$ asmtprmrydsbltycd : num [1:3627] 0 10 40 82 10 80 50 10 50 82 ...## ..$ asmtscndrydsbltycd : num [1:3627] 0 0 20 0 0 80 0 0 0 0 ...## ..$ Entry : num [1:3627] 123 88 105 153 437 307 305 42 59 304 ...## ..$ Theta : num [1:3627] 1.27 1.55 3.28 4.48 2.67 ...## ..$ Status : num [1:3627] 1 1 1 1 1 0 1 1 1 0 ...## ..$ Count : num [1:3627] 36 36 36 36 36 36 36 36 36 36 ...## ..$ RawScore : num [1:3627] 23 25 33 35 31 36 34 18 3 36 ...## ..$ SE : num [1:3627] 0.371 0.385 0.619 1.023 0.501 ...## ..$ Infit : num [1:3627] 0.93 0.95 0.9 0.93 0.92 1 1.06 1.55 0.85 1 ...## ..$ Infit_Z : num [1:3627] -0.34 -0.37 -0.04 0.23 -0.18 0 0.31 2.56 0.06 0 ...## ..$ Outfit : num [1:3627] 0.82 0.81 1.63 0.35 0.88 1 0.86 1.74 0.17 1 ...## ..$ Outfit_Z : num [1:3627] -0.62 -0.56 1.03 -0.16 -0.12 0 0.17 3.31 -0.37 0 ...## ..$ Displacement : num [1:3627] 0.0018 0.0019 0.0022 0.0023 0.0021 0.0024 0.0022 0.0017 0.0009 0.0024 ...## ..$ PointMeasureCorr : num [1:3627] 0.42 0.42 0.3 0.27 0.31 0 0.14 -0.12 0.32 0 ...## ..$ Weight : num [1:3627] 1 1 1 1 1 1 1 1 1 1 ...## ..$ ObservMatch : num [1:3627] 75 80.6 91.7 97.2 86.1 100 94.4 50 97.2 100 ...## ..$ ExpectMatch : num [1:3627] 68.3 72 91.7 97.2 86.1 100 94.4 65 97.2 100 ...## ..$ PointMeasureExpected: num [1:3627] 0.35 0.33 0.2 0.12 0.25 0 0.17 0.38 0.17 0 ...## ..$ RMSR : num [1:3627] 0.42 0.39 0.26 0.16 0.31 0 0.23 0.51 0.14 0 ...## ..$ WMLE : num [1:3627] 1.25 1.68 3.13 4 2.59 ...## $ Math : tibble[,25] [3,629 × 25] (S3: tbl_df/tbl/data.frame)## ..$ ssid : num [1:3629] 10129634 10926496 10616063 10139443 8887381 ...## ..$ grade : int [1:3629] 11 11 11 11 11 11 11 11 11 11 ...## ..$ year : int [1:3629] 11 11 11 11 11 11 11 11 11 11 ...## ..$ content : chr [1:3629] "Math" "Math" "Math" "Math" ...## ..$ testeventid : num [1:3629] 147564 151249 146976 151229 147676 ...## ..$ asmtprmrydsbltycd : num [1:3629] 10 80 10 10 80 82 10 10 40 82 ...## ..$ asmtscndrydsbltycd : num [1:3629] 0 90 0 0 10 80 50 0 50 0 ...## ..$ Entry : num [1:3629] 161 344 321 167 101 131 141 357 14 367 ...## ..$ Theta : num [1:3629] 0.514 1.965 0.899 1.965 -4.724 ...## ..$ Status : num [1:3629] 1 1 1 1 -1 1 1 1 1 1 ...## ..$ Count : num [1:3629] 36 36 36 36 36 36 36 36 36 36 ...## ..$ RawScore : num [1:3629] 19 29 22 29 0 14 18 25 19 22 ...## ..$ SE : num [1:3629] 0.356 0.436 0.362 0.436 1.839 ...## ..$ Infit : num [1:3629] 0.97 1.03 0.99 1 1 1.01 0.99 0.85 0.96 1.06 ...## ..$ Infit_Z : num [1:3629] -0.18 0.26 -0.09 0.1 0 0.13 -0.06 -0.81 -0.39 0.5 ...## ..$ Outfit : num [1:3629] 0.97 1.03 1.01 0.87 1 0.93 1 0.77 0.97 0.99 ...## ..$ Outfit_Z : num [1:3629] -0.17 0.23 0.17 -0.46 0 -0.03 0.03 -0.87 -0.19 0.07 ...## ..$ Displacement : num [1:3629] 5e-04 5e-04 5e-04 5e-04 -7e-04 4e-04 5e-04 5e-04 5e-04 5e-04 ...## ..$ PointMeasureCorr : num [1:3629] 0.38 0.28 0.34 0.29 0 0.34 0.38 0.45 0.38 0.27 ...## ..$ Weight : num [1:3629] 1 1 1 1 1 1 1 1 1 1 ...## ..$ ObservMatch : num [1:3629] 66.7 80.6 66.7 80.6 100 61.1 69.4 75 63.9 61.1 ...## ..$ ExpectMatch : num [1:3629] 64.4 80.5 66 80.5 100 68.3 64.6 70.3 64.4 66 ...## ..$ PointMeasureExpected: num [1:3629] 0.35 0.25 0.33 0.25 0 0.35 0.35 0.3 0.35 0.33 ...## ..$ RMSR : num [1:3629] 0.46 0.39 0.45 0.39 0 0.46 0.46 0.4 0.46 0.48 ...## ..$ WMLE : num [1:3629] 0.512 1.915 0.888 1.915 -4.724 ...## $ Rdg : tibble[,25] [3,627 × 25] (S3: tbl_df/tbl/data.frame)## ..$ ssid : num [1:3627] 18631185 18342736 10897771 7663935 6709613 ...## ..$ grade : int [1:3627] 11 11 11 11 11 11 11 11 11 11 ...## ..$ year : int [1:3627] 11 11 11 11 11 11 11 11 11 11 ...## ..$ content : chr [1:3627] "Rdg" "Rdg" "Rdg" "Rdg" ...## ..$ testeventid : num [1:3627] 145641 147717 145196 149014 146545 ...## ..$ asmtprmrydsbltycd : num [1:3627] 10 90 10 80 0 10 10 80 82 82 ...## ..$ asmtscndrydsbltycd : num [1:3627] 0 0 0 10 0 82 0 70 0 0 ...## ..$ Entry : num [1:3627] 444 444 343 74 3 77 181 261 297 182 ...## ..$ Theta : num [1:3627] 2.24 3.48 2.72 -2.72 0.12 -1.39 -0.67 4.73 4.73 4.73 ...## ..$ Status : num [1:3627] 1 1 1 1 1 1 1 0 0 0 ...## ..$ Count : num [1:3627] 19 19 19 19 19 19 19 19 19 19 ...## ..$ RawScore : num [1:3627] 16 18 17 1 8 3 5 19 19 19 ...## ..$ SE : num [1:3627] 0.64 1.03 0.76 1.06 0.49 0.66 0.55 1.84 1.84 1.84 ...## ..$ Infit : num [1:3627] 1.07 1.02 1.1 1 0.98 0.73 1.08 1 1 1 ...## ..$ Infit_Z : num [1:3627] 0.29 0.31 0.53 0.29 -0.11 -0.54 0.37 0 0 0 ...## ..$ Outfit : num [1:3627] 1.17 0.94 1.22 0.76 0.95 0.62 1.09 1 1 1 ...## ..$ Outfit_Z : num [1:3627] 0.45 0.32 0.71 0 -0.22 -0.62 0.42 0 0 0 ...## ..$ Displacement : num [1:3627] 0 0 0 0 0 0 0 0 0 0 ...## ..$ PointMeasureCorr : num [1:3627] 0.01 0.08 0.05 0 0.32 0.68 0.2 0 0 0 ...## ..$ Weight : num [1:3627] 1 1 1 1 1 1 1 1 1 1 ...## ..$ ObservMatch : num [1:3627] 84.2 94.7 89.5 94.7 63.2 84.2 78.9 100 100 100 ...## ..$ ExpectMatch : num [1:3627] 84.2 94.7 89.5 94.7 64.2 85.1 76.3 100 100 100 ...## ..$ PointMeasureExpected: num [1:3627] 0.17 0.1 0.14 0.24 0.3 0.31 0.32 0 0 0 ...## ..$ RMSR : num [1:3627] 0.38 0.22 0.31 0.22 0.47 0.34 0.36 0 0 0 ...## ..$ WMLE : num [1:3627] 2.11 3.02 2.5 -2.27 0.14 -0.9 -0.6 4.73 4.73 4.73 ...## $ Science: tibble[,25] [1,435 × 25] (S3: tbl_df/tbl/data.frame)## ..$ ssid : num [1:1435] 7617607 7642717 10341706 9811494 10469745 ...## ..$ grade : int [1:1435] 11 11 11 11 11 11 11 11 11 11 ...## ..$ year : int [1:1435] 11 11 11 11 11 11 11 11 11 11 ...## ..$ content : chr [1:1435] "Science" "Science" "Science" "Science" ...## ..$ testeventid : num [1:1435] 148838 144146 149634 146456 144426 ...## ..$ asmtprmrydsbltycd : num [1:1435] 10 82 80 10 90 10 10 80 80 10 ...## ..$ asmtscndrydsbltycd : num [1:1435] 0 80 20 0 50 80 70 0 50 0 ...## ..$ Entry : num [1:1435] 28 43 274 118 291 91 58 130 297 337 ...## ..$ Theta : num [1:1435] 0.875 2.647 4.17 5.404 5.404 ...## ..$ Status : num [1:1435] 1 1 1 0 0 1 1 1 1 1 ...## ..$ Count : num [1:1435] 36 36 36 36 36 36 36 36 36 36 ...## ..$ RawScore : num [1:1435] 22 32 35 36 36 26 2 34 34 28 ...## ..$ SE : num [1:1435] 0.361 0.544 1.021 1.835 1.835 ...## ..$ Infit : num [1:1435] 1.3 0.94 1.05 1.04 0.99 0.9 0.97 0.99 0.98 0.9 ...## ..$ Infit_Z : num [1:1435] 2.35 0.02 0.37 0.35 0 -0.62 0.17 0.15 0.16 -0.52 ...## ..$ Outfit : num [1:1435] 1.22 0.76 1.5 1.2 0.48 0.85 0.69 1.57 0.67 0.8 ...## ..$ Outfit_Z : num [1:1435] 1.87 -0.27 0.91 0.6 -0.16 -0.87 -0.41 0.74 0.01 -0.76 ...## ..$ Displacement : num [1:1435] 0.0011 0.0016 0.0016 0.0027 0.0027 0.0013 0.0007 0.0016 0.0016 0.0014 ...## ..$ PointMeasureCorr : num [1:1435] 0.07 0.3 -0.07 0 0.29 0.46 0.28 0.05 0.22 0.45 ...## ..$ Weight : num [1:1435] 1 1 1 1 1 1 1 1 1 1 ...## ..$ ObservMatch : num [1:1435] 55.6 88.9 97.2 100 100 75 94.4 94.4 94.4 80.6 ...## ..$ ExpectMatch : num [1:1435] 66.1 88.9 97.2 100 100 73.2 94.4 94.4 94.4 77.9 ...## ..$ PointMeasureExpected: num [1:1435] 0.32 0.22 0.12 0 0 0.3 0.15 0.16 0.16 0.28 ...## ..$ RMSR : num [1:1435] 0.52 0.3 0.17 0 0 0.39 0.22 0.22 0.22 0.36 ...## ..$ WMLE : num [1:1435] 0.863 2.543 3.692 5.404 5.404 ...## $ Wri : tibble[,25] [3,627 × 25] (S3: tbl_df/tbl/data.frame)## ..$ ssid : num [1:3627] 7653093 7640498 7650957 9305807 18060455 ...## ..$ grade : int [1:3627] 11 11 11 11 11 11 11 11 11 11 ...## ..$ year : int [1:3627] 11 11 11 11 11 11 11 11 11 11 ...## ..$ content : chr [1:3627] "Wri" "Wri" "Wri" "Wri" ...## ..$ testeventid : num [1:3627] 148893 148868 144397 148988 144865 ...## ..$ asmtprmrydsbltycd : num [1:3627] 10 10 90 0 82 10 80 0 10 82 ...## ..$ asmtscndrydsbltycd : num [1:3627] 0 0 80 0 0 0 50 0 0 10 ...## ..$ Entry : num [1:3627] 61 48 63 107 443 214 267 344 423 27 ...## ..$ Theta : num [1:3627] 1.67 2.17 -1.56 5.01 2.17 0.38 0.79 5.01 -0.48 2.78 ...## ..$ Status : num [1:3627] 1 1 1 0 1 1 1 0 1 1 ...## ..$ Count : num [1:3627] 13 13 13 13 13 13 13 13 13 13 ...## ..$ RawScore : num [1:3627] 9 10 2 13 10 6 7 13 4 11 ...## ..$ SE : num [1:3627] 0.69 0.74 0.83 1.87 0.74 0.64 0.65 1.87 0.68 0.84 ...## ..$ Infit : num [1:3627] 0.52 0.65 0.61 1 1.01 0.46 0.47 1 0.9 0.88 ...## ..$ Infit_Z : num [1:3627] -2.07 -1.02 -0.79 0 0.24 -2.3 -2.2 0 -0.16 -0.21 ...## ..$ Outfit : num [1:3627] 0.44 0.52 0.33 1 0.74 0.41 0.4 1 1.32 0.54 ...## ..$ Outfit_Z : num [1:3627] -1.65 -0.81 -0.47 0 -0.06 -1.88 -1.85 0 -0.14 -0.16 ...## ..$ Displacement : num [1:3627] 0 0 0 0 0 0 0 0 0 0 ...## ..$ PointMeasureCorr : num [1:3627] 0.86 0.73 0.62 0 0.51 0.87 0.87 0 0.56 0.56 ...## ..$ Weight : num [1:3627] 1 1 1 1 1 1 1 1 1 1 ...## ..$ ObservMatch : num [1:3627] 100 92.3 84.6 100 76.9 100 100 100 92.3 84.6 ...## ..$ ExpectMatch : num [1:3627] 75.5 79.7 84.6 100 79.7 72.9 73 100 74.5 84.6 ...## ..$ PointMeasureExpected: num [1:3627] 0.48 0.44 0.34 0 0.44 0.5 0.51 0 0.45 0.38 ...## ..$ RMSR : num [1:3627] 0.31 0.34 0.26 0 0.38 0.29 0.29 0 0.37 0.31 ...## ..$ WMLE : num [1:3627] 1.61 2.08 -1.38 5.01 2.08 0.38 0.78 5.01 -0.43 2.61 ...53 / 62
We could use this method
m1 <- map( splt_content, ~lm(Theta ~ asmtprmrydsbltycd, data = .x))m2 <- map( splt_content, ~lm(Theta ~ asmtprmrydsbltycd + asmtscndrydsbltycd, data = .x))m3 <- map( splt_content, ~lm(Theta ~ asmtprmrydsbltycd * asmtscndrydsbltycd, data = .x))- We could then go through and conduct tests to see which model had better fit indices, etc.
54 / 62
Alternative
- Create a data frame with a list column
d %>% nest(-content)## # A tibble: 5 x 2## content data ## <chr> <list> ## 1 ELA <tibble[,24] [3,627 × 24]>## 2 Math <tibble[,24] [3,629 × 24]>## 3 Rdg <tibble[,24] [3,627 × 24]>## 4 Science <tibble[,24] [1,435 × 24]>## 5 Wri <tibble[,24] [3,627 × 24]>55 / 62
mods## # A tibble: 5 x 5## content data m1 m2 m3 ## <chr> <list> <list> <list> <list>## 1 ELA <tibble[,24] [3,627 × 24]> <lm> <lm> <lm> ## 2 Math <tibble[,24] [3,629 × 24]> <lm> <lm> <lm> ## 3 Rdg <tibble[,24] [3,627 × 24]> <lm> <lm> <lm> ## 4 Science <tibble[,24] [1,435 × 24]> <lm> <lm> <lm> ## 5 Wri <tibble[,24] [3,627 × 24]> <lm> <lm> <lm>57 / 62
Part of the benefit
It's a normal data frame!
mods %>% pivot_longer( m1:m3, names_to = "model", values_to = "output" )## # A tibble: 15 x 4## content data model output## <chr> <list> <chr> <list>## 1 ELA <tibble[,24] [3,627 × 24]> m1 <lm> ## 2 ELA <tibble[,24] [3,627 × 24]> m2 <lm> ## 3 ELA <tibble[,24] [3,627 × 24]> m3 <lm> ## 4 Math <tibble[,24] [3,629 × 24]> m1 <lm> ## 5 Math <tibble[,24] [3,629 × 24]> m2 <lm> ## 6 Math <tibble[,24] [3,629 × 24]> m3 <lm> ## 7 Rdg <tibble[,24] [3,627 × 24]> m1 <lm> ## 8 Rdg <tibble[,24] [3,627 × 24]> m2 <lm> ## 9 Rdg <tibble[,24] [3,627 × 24]> m3 <lm> ## 10 Science <tibble[,24] [1,435 × 24]> m1 <lm> ## 11 Science <tibble[,24] [1,435 × 24]> m2 <lm> ## 12 Science <tibble[,24] [1,435 × 24]> m3 <lm> ## 13 Wri <tibble[,24] [3,627 × 24]> m1 <lm> ## 14 Wri <tibble[,24] [3,627 × 24]> m2 <lm> ## 15 Wri <tibble[,24] [3,627 × 24]> m3 <lm>58 / 62
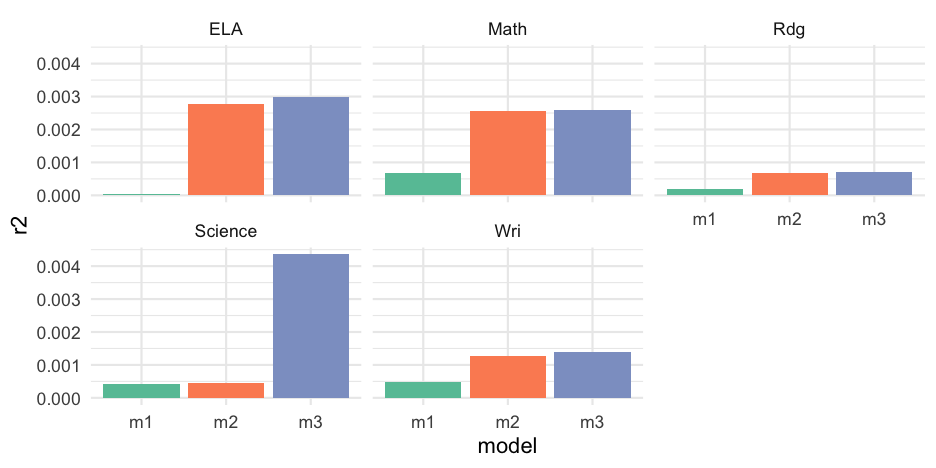
Extract all R2
r2 <- mods %>% pivot_longer( m1:m3, names_to = "model", values_to = "output" ) %>% mutate(r2 = map_dbl(output, ~summary(.x)$r.squared))r2## # A tibble: 15 x 5## content data model output r2## <chr> <list> <chr> <list> <dbl>## 1 ELA <tibble[,24] [3,627 × 24]> m1 <lm> 0.00002742625## 2 ELA <tibble[,24] [3,627 × 24]> m2 <lm> 0.002784211 ## 3 ELA <tibble[,24] [3,627 × 24]> m3 <lm> 0.002994548 ## 4 Math <tibble[,24] [3,629 × 24]> m1 <lm> 0.0006718361 ## 5 Math <tibble[,24] [3,629 × 24]> m2 <lm> 0.002575408 ## 6 Math <tibble[,24] [3,629 × 24]> m3 <lm> 0.002586228 ## 7 Rdg <tibble[,24] [3,627 × 24]> m1 <lm> 0.0001925962 ## 8 Rdg <tibble[,24] [3,627 × 24]> m2 <lm> 0.0006773540 ## 9 Rdg <tibble[,24] [3,627 × 24]> m3 <lm> 0.0007050212 ## 10 Science <tibble[,24] [1,435 × 24]> m1 <lm> 0.0004085780 ## 11 Science <tibble[,24] [1,435 × 24]> m2 <lm> 0.0004520424 ## 12 Science <tibble[,24] [1,435 × 24]> m3 <lm> 0.004354060 ## 13 Wri <tibble[,24] [3,627 × 24]> m1 <lm> 0.0004902093 ## 14 Wri <tibble[,24] [3,627 × 24]> m2 <lm> 0.001256362 ## 15 Wri <tibble[,24] [3,627 × 24]> m3 <lm> 0.00139198059 / 62