List columns
Daniel Anderson
Week 4, Class 2
Parse file data
d <- d %>% mutate( file = str_replace_all( file, here::here("data", "pfiles_sim"), "" ), grade = str_replace_all(file, "/g(\\d?\\d).+", "\\1"), grade = as.integer(grade), year = str_replace_all( file, ".+files(\\d\\d)_sim.+", "\\1" ), year = as.integer(year), content = str_replace_all( file, "/g\\d?\\d(.+)pfiles.+", "\\1" ))Data pre-processing
- The disability variables are stored as numbers, we need them as factors
- We'll make the names easier in the process
d <- d %>% mutate(primary = as.factor(asmtprmrydsbltycd), secondary = as.factor(asmtscndrydsbltycd))If you're interested in what the specific codes refer to, see here.
Split the data
The base method we've been using...
splt_content <- split(d, d$content)str(splt_content)## List of 5## $ ELA : tibble[,27] [3,627 × 27] (S3: tbl_df/tbl/data.frame)## ..$ ssid : num [1:3627] 9466908 7683685 9025693 10099824 18886078 ...## ..$ grade : int [1:3627] 11 11 11 11 11 11 11 11 11 11 ...## ..$ year : int [1:3627] 18 18 18 18 18 18 18 18 18 18 ...## ..$ content : chr [1:3627] "ELA" "ELA" "ELA" "ELA" ...## ..$ testeventid : num [1:3627] 148933 147875 143699 143962 150680 ...## ..$ asmtprmrydsbltycd : num [1:3627] 0 10 40 82 10 80 50 10 50 82 ...## ..$ asmtscndrydsbltycd : num [1:3627] 0 0 20 0 0 80 0 0 0 0 ...## ..$ Entry : num [1:3627] 123 88 105 153 437 307 305 42 59 304 ...## ..$ Theta : num [1:3627] 1.27 1.55 3.28 4.48 2.67 ...## ..$ Status : num [1:3627] 1 1 1 1 1 0 1 1 1 0 ...## ..$ Count : num [1:3627] 36 36 36 36 36 36 36 36 36 36 ...## ..$ RawScore : num [1:3627] 23 25 33 35 31 36 34 18 3 36 ...## ..$ SE : num [1:3627] 0.371 0.385 0.619 1.023 0.501 ...## ..$ Infit : num [1:3627] 0.93 0.95 0.9 0.93 0.92 1 1.06 1.55 0.85 1 ...## ..$ Infit_Z : num [1:3627] -0.34 -0.37 -0.04 0.23 -0.18 0 0.31 2.56 0.06 0 ...## ..$ Outfit : num [1:3627] 0.82 0.81 1.63 0.35 0.88 1 0.86 1.74 0.17 1 ...## ..$ Outfit_Z : num [1:3627] -0.62 -0.56 1.03 -0.16 -0.12 0 0.17 3.31 -0.37 0 ...## ..$ Displacement : num [1:3627] 0.0018 0.0019 0.0022 0.0023 0.0021 0.0024 0.0022 0.0017 0.0009 0.0024 ...## ..$ PointMeasureCorr : num [1:3627] 0.42 0.42 0.3 0.27 0.31 0 0.14 -0.12 0.32 0 ...## ..$ Weight : num [1:3627] 1 1 1 1 1 1 1 1 1 1 ...## ..$ ObservMatch : num [1:3627] 75 80.6 91.7 97.2 86.1 100 94.4 50 97.2 100 ...## ..$ ExpectMatch : num [1:3627] 68.3 72 91.7 97.2 86.1 100 94.4 65 97.2 100 ...## ..$ PointMeasureExpected: num [1:3627] 0.35 0.33 0.2 0.12 0.25 0 0.17 0.38 0.17 0 ...## ..$ RMSR : num [1:3627] 0.42 0.39 0.26 0.16 0.31 0 0.23 0.51 0.14 0 ...## ..$ WMLE : num [1:3627] 1.25 1.68 3.13 4 2.59 ...## ..$ primary : Factor w/ 12 levels "0","10","20",..: 1 2 4 11 2 10 6 2 6 11 ...## ..$ secondary : Factor w/ 12 levels "0","10","20",..: 1 1 3 1 1 10 1 1 1 1 ...## $ Math : tibble[,27] [3,629 × 27] (S3: tbl_df/tbl/data.frame)## ..$ ssid : num [1:3629] 10129634 10926496 10616063 10139443 8887381 ...## ..$ grade : int [1:3629] 11 11 11 11 11 11 11 11 11 11 ...## ..$ year : int [1:3629] 18 18 18 18 18 18 18 18 18 18 ...## ..$ content : chr [1:3629] "Math" "Math" "Math" "Math" ...## ..$ testeventid : num [1:3629] 147564 151249 146976 151229 147676 ...## ..$ asmtprmrydsbltycd : num [1:3629] 10 80 10 10 80 82 10 10 40 82 ...## ..$ asmtscndrydsbltycd : num [1:3629] 0 90 0 0 10 80 50 0 50 0 ...## ..$ Entry : num [1:3629] 161 344 321 167 101 131 141 357 14 367 ...## ..$ Theta : num [1:3629] 0.514 1.965 0.899 1.965 -4.724 ...## ..$ Status : num [1:3629] 1 1 1 1 -1 1 1 1 1 1 ...## ..$ Count : num [1:3629] 36 36 36 36 36 36 36 36 36 36 ...## ..$ RawScore : num [1:3629] 19 29 22 29 0 14 18 25 19 22 ...## ..$ SE : num [1:3629] 0.356 0.436 0.362 0.436 1.839 ...## ..$ Infit : num [1:3629] 0.97 1.03 0.99 1 1 1.01 0.99 0.85 0.96 1.06 ...## ..$ Infit_Z : num [1:3629] -0.18 0.26 -0.09 0.1 0 0.13 -0.06 -0.81 -0.39 0.5 ...## ..$ Outfit : num [1:3629] 0.97 1.03 1.01 0.87 1 0.93 1 0.77 0.97 0.99 ...## ..$ Outfit_Z : num [1:3629] -0.17 0.23 0.17 -0.46 0 -0.03 0.03 -0.87 -0.19 0.07 ...## ..$ Displacement : num [1:3629] 5e-04 5e-04 5e-04 5e-04 -7e-04 4e-04 5e-04 5e-04 5e-04 5e-04 ...## ..$ PointMeasureCorr : num [1:3629] 0.38 0.28 0.34 0.29 0 0.34 0.38 0.45 0.38 0.27 ...## ..$ Weight : num [1:3629] 1 1 1 1 1 1 1 1 1 1 ...## ..$ ObservMatch : num [1:3629] 66.7 80.6 66.7 80.6 100 61.1 69.4 75 63.9 61.1 ...## ..$ ExpectMatch : num [1:3629] 64.4 80.5 66 80.5 100 68.3 64.6 70.3 64.4 66 ...## ..$ PointMeasureExpected: num [1:3629] 0.35 0.25 0.33 0.25 0 0.35 0.35 0.3 0.35 0.33 ...## ..$ RMSR : num [1:3629] 0.46 0.39 0.45 0.39 0 0.46 0.46 0.4 0.46 0.48 ...## ..$ WMLE : num [1:3629] 0.512 1.915 0.888 1.915 -4.724 ...## ..$ primary : Factor w/ 12 levels "0","10","20",..: 2 10 2 2 10 11 2 2 4 11 ...## ..$ secondary : Factor w/ 12 levels "0","10","20",..: 1 12 1 1 2 10 6 1 6 1 ...## $ Rdg : tibble[,27] [3,627 × 27] (S3: tbl_df/tbl/data.frame)## ..$ ssid : num [1:3627] 18631185 18342736 10897771 7663935 6709613 ...## ..$ grade : int [1:3627] 11 11 11 11 11 11 11 11 11 11 ...## ..$ year : int [1:3627] 18 18 18 18 18 18 18 18 18 18 ...## ..$ content : chr [1:3627] "Rdg" "Rdg" "Rdg" "Rdg" ...## ..$ testeventid : num [1:3627] 145641 147717 145196 149014 146545 ...## ..$ asmtprmrydsbltycd : num [1:3627] 10 90 10 80 0 10 10 80 82 82 ...## ..$ asmtscndrydsbltycd : num [1:3627] 0 0 0 10 0 82 0 70 0 0 ...## ..$ Entry : num [1:3627] 444 444 343 74 3 77 181 261 297 182 ...## ..$ Theta : num [1:3627] 2.24 3.48 2.72 -2.72 0.12 -1.39 -0.67 4.73 4.73 4.73 ...## ..$ Status : num [1:3627] 1 1 1 1 1 1 1 0 0 0 ...## ..$ Count : num [1:3627] 19 19 19 19 19 19 19 19 19 19 ...## ..$ RawScore : num [1:3627] 16 18 17 1 8 3 5 19 19 19 ...## ..$ SE : num [1:3627] 0.64 1.03 0.76 1.06 0.49 0.66 0.55 1.84 1.84 1.84 ...## ..$ Infit : num [1:3627] 1.07 1.02 1.1 1 0.98 0.73 1.08 1 1 1 ...## ..$ Infit_Z : num [1:3627] 0.29 0.31 0.53 0.29 -0.11 -0.54 0.37 0 0 0 ...## ..$ Outfit : num [1:3627] 1.17 0.94 1.22 0.76 0.95 0.62 1.09 1 1 1 ...## ..$ Outfit_Z : num [1:3627] 0.45 0.32 0.71 0 -0.22 -0.62 0.42 0 0 0 ...## ..$ Displacement : num [1:3627] 0 0 0 0 0 0 0 0 0 0 ...## ..$ PointMeasureCorr : num [1:3627] 0.01 0.08 0.05 0 0.32 0.68 0.2 0 0 0 ...## ..$ Weight : num [1:3627] 1 1 1 1 1 1 1 1 1 1 ...## ..$ ObservMatch : num [1:3627] 84.2 94.7 89.5 94.7 63.2 84.2 78.9 100 100 100 ...## ..$ ExpectMatch : num [1:3627] 84.2 94.7 89.5 94.7 64.2 85.1 76.3 100 100 100 ...## ..$ PointMeasureExpected: num [1:3627] 0.17 0.1 0.14 0.24 0.3 0.31 0.32 0 0 0 ...## ..$ RMSR : num [1:3627] 0.38 0.22 0.31 0.22 0.47 0.34 0.36 0 0 0 ...## ..$ WMLE : num [1:3627] 2.11 3.02 2.5 -2.27 0.14 -0.9 -0.6 4.73 4.73 4.73 ...## ..$ primary : Factor w/ 12 levels "0","10","20",..: 2 12 2 10 1 2 2 10 11 11 ...## ..$ secondary : Factor w/ 12 levels "0","10","20",..: 1 1 1 2 1 11 1 8 1 1 ...## $ Science: tibble[,27] [1,435 × 27] (S3: tbl_df/tbl/data.frame)## ..$ ssid : num [1:1435] 7617607 7642717 10341706 9811494 10469745 ...## ..$ grade : int [1:1435] 11 11 11 11 11 11 11 11 11 11 ...## ..$ year : int [1:1435] 18 18 18 18 18 18 18 18 18 18 ...## ..$ content : chr [1:1435] "Science" "Science" "Science" "Science" ...## ..$ testeventid : num [1:1435] 148838 144146 149634 146456 144426 ...## ..$ asmtprmrydsbltycd : num [1:1435] 10 82 80 10 90 10 10 80 80 10 ...## ..$ asmtscndrydsbltycd : num [1:1435] 0 80 20 0 50 80 70 0 50 0 ...## ..$ Entry : num [1:1435] 28 43 274 118 291 91 58 130 297 337 ...## ..$ Theta : num [1:1435] 0.875 2.647 4.17 5.404 5.404 ...## ..$ Status : num [1:1435] 1 1 1 0 0 1 1 1 1 1 ...## ..$ Count : num [1:1435] 36 36 36 36 36 36 36 36 36 36 ...## ..$ RawScore : num [1:1435] 22 32 35 36 36 26 2 34 34 28 ...## ..$ SE : num [1:1435] 0.361 0.544 1.021 1.835 1.835 ...## ..$ Infit : num [1:1435] 1.3 0.94 1.05 1.04 0.99 0.9 0.97 0.99 0.98 0.9 ...## ..$ Infit_Z : num [1:1435] 2.35 0.02 0.37 0.35 0 -0.62 0.17 0.15 0.16 -0.52 ...## ..$ Outfit : num [1:1435] 1.22 0.76 1.5 1.2 0.48 0.85 0.69 1.57 0.67 0.8 ...## ..$ Outfit_Z : num [1:1435] 1.87 -0.27 0.91 0.6 -0.16 -0.87 -0.41 0.74 0.01 -0.76 ...## ..$ Displacement : num [1:1435] 0.0011 0.0016 0.0016 0.0027 0.0027 0.0013 0.0007 0.0016 0.0016 0.0014 ...## ..$ PointMeasureCorr : num [1:1435] 0.07 0.3 -0.07 0 0.29 0.46 0.28 0.05 0.22 0.45 ...## ..$ Weight : num [1:1435] 1 1 1 1 1 1 1 1 1 1 ...## ..$ ObservMatch : num [1:1435] 55.6 88.9 97.2 100 100 75 94.4 94.4 94.4 80.6 ...## ..$ ExpectMatch : num [1:1435] 66.1 88.9 97.2 100 100 73.2 94.4 94.4 94.4 77.9 ...## ..$ PointMeasureExpected: num [1:1435] 0.32 0.22 0.12 0 0 0.3 0.15 0.16 0.16 0.28 ...## ..$ RMSR : num [1:1435] 0.52 0.3 0.17 0 0 0.39 0.22 0.22 0.22 0.36 ...## ..$ WMLE : num [1:1435] 0.863 2.543 3.692 5.404 5.404 ...## ..$ primary : Factor w/ 12 levels "0","10","20",..: 2 11 10 2 12 2 2 10 10 2 ...## ..$ secondary : Factor w/ 12 levels "0","10","20",..: 1 10 3 1 6 10 8 1 6 1 ...## $ Wri : tibble[,27] [3,627 × 27] (S3: tbl_df/tbl/data.frame)## ..$ ssid : num [1:3627] 7653093 7640498 7650957 9305807 18060455 ...## ..$ grade : int [1:3627] 11 11 11 11 11 11 11 11 11 11 ...## ..$ year : int [1:3627] 18 18 18 18 18 18 18 18 18 18 ...## ..$ content : chr [1:3627] "Wri" "Wri" "Wri" "Wri" ...## ..$ testeventid : num [1:3627] 148893 148868 144397 148988 144865 ...## ..$ asmtprmrydsbltycd : num [1:3627] 10 10 90 0 82 10 80 0 10 82 ...## ..$ asmtscndrydsbltycd : num [1:3627] 0 0 80 0 0 0 50 0 0 10 ...## ..$ Entry : num [1:3627] 61 48 63 107 443 214 267 344 423 27 ...## ..$ Theta : num [1:3627] 1.67 2.17 -1.56 5.01 2.17 0.38 0.79 5.01 -0.48 2.78 ...## ..$ Status : num [1:3627] 1 1 1 0 1 1 1 0 1 1 ...## ..$ Count : num [1:3627] 13 13 13 13 13 13 13 13 13 13 ...## ..$ RawScore : num [1:3627] 9 10 2 13 10 6 7 13 4 11 ...## ..$ SE : num [1:3627] 0.69 0.74 0.83 1.87 0.74 0.64 0.65 1.87 0.68 0.84 ...## ..$ Infit : num [1:3627] 0.52 0.65 0.61 1 1.01 0.46 0.47 1 0.9 0.88 ...## ..$ Infit_Z : num [1:3627] -2.07 -1.02 -0.79 0 0.24 -2.3 -2.2 0 -0.16 -0.21 ...## ..$ Outfit : num [1:3627] 0.44 0.52 0.33 1 0.74 0.41 0.4 1 1.32 0.54 ...## ..$ Outfit_Z : num [1:3627] -1.65 -0.81 -0.47 0 -0.06 -1.88 -1.85 0 -0.14 -0.16 ...## ..$ Displacement : num [1:3627] 0 0 0 0 0 0 0 0 0 0 ...## ..$ PointMeasureCorr : num [1:3627] 0.86 0.73 0.62 0 0.51 0.87 0.87 0 0.56 0.56 ...## ..$ Weight : num [1:3627] 1 1 1 1 1 1 1 1 1 1 ...## ..$ ObservMatch : num [1:3627] 100 92.3 84.6 100 76.9 100 100 100 92.3 84.6 ...## ..$ ExpectMatch : num [1:3627] 75.5 79.7 84.6 100 79.7 72.9 73 100 74.5 84.6 ...## ..$ PointMeasureExpected: num [1:3627] 0.48 0.44 0.34 0 0.44 0.5 0.51 0 0.45 0.38 ...## ..$ RMSR : num [1:3627] 0.31 0.34 0.26 0 0.38 0.29 0.29 0 0.37 0.31 ...## ..$ WMLE : num [1:3627] 1.61 2.08 -1.38 5.01 2.08 0.38 0.78 5.01 -0.43 2.61 ...## ..$ primary : Factor w/ 12 levels "0","10","20",..: 2 2 12 1 11 2 10 1 2 11 ...## ..$ secondary : Factor w/ 12 levels "0","10","20",..: 1 1 10 1 1 1 6 1 1 2 ...We could use this method
m1 <- map( splt_content, ~lm(Theta ~ asmtprmrydsbltycd, data = .x))m2 <- map( splt_content, ~lm(Theta ~ asmtprmrydsbltycd + asmtscndrydsbltycd, data = .x))m3 <- map( splt_content, ~lm(Theta ~ asmtprmrydsbltycd * asmtscndrydsbltycd, data = .x))- Then conduct tests to see which model fit better, etc.
Alternative
- Create a data frame with a list column
by_content <- d %>% group_by(content) %>% nest()by_content## # A tibble: 5 x 2## # Groups: content [5]## content data ## <chr> <list> ## 1 ELA <tibble[,26] [3,627 × 26]>## 2 Math <tibble[,26] [3,629 × 26]>## 3 Rdg <tibble[,26] [3,627 × 26]>## 4 Science <tibble[,26] [1,435 × 26]>## 5 Wri <tibble[,26] [3,627 × 26]>What's going on here?
str(by_content$data)## List of 5## $ : tibble[,26] [3,627 × 26] (S3: tbl_df/tbl/data.frame)## ..$ ssid : num [1:3627] 9466908 7683685 9025693 10099824 18886078 ...## ..$ grade : int [1:3627] 11 11 11 11 11 11 11 11 11 11 ...## ..$ year : int [1:3627] 18 18 18 18 18 18 18 18 18 18 ...## ..$ testeventid : num [1:3627] 148933 147875 143699 143962 150680 ...## ..$ asmtprmrydsbltycd : num [1:3627] 0 10 40 82 10 80 50 10 50 82 ...## ..$ asmtscndrydsbltycd : num [1:3627] 0 0 20 0 0 80 0 0 0 0 ...## ..$ Entry : num [1:3627] 123 88 105 153 437 307 305 42 59 304 ...## ..$ Theta : num [1:3627] 1.27 1.55 3.28 4.48 2.67 ...## ..$ Status : num [1:3627] 1 1 1 1 1 0 1 1 1 0 ...## ..$ Count : num [1:3627] 36 36 36 36 36 36 36 36 36 36 ...## ..$ RawScore : num [1:3627] 23 25 33 35 31 36 34 18 3 36 ...## ..$ SE : num [1:3627] 0.371 0.385 0.619 1.023 0.501 ...## ..$ Infit : num [1:3627] 0.93 0.95 0.9 0.93 0.92 1 1.06 1.55 0.85 1 ...## ..$ Infit_Z : num [1:3627] -0.34 -0.37 -0.04 0.23 -0.18 0 0.31 2.56 0.06 0 ...## ..$ Outfit : num [1:3627] 0.82 0.81 1.63 0.35 0.88 1 0.86 1.74 0.17 1 ...## ..$ Outfit_Z : num [1:3627] -0.62 -0.56 1.03 -0.16 -0.12 0 0.17 3.31 -0.37 0 ...## ..$ Displacement : num [1:3627] 0.0018 0.0019 0.0022 0.0023 0.0021 0.0024 0.0022 0.0017 0.0009 0.0024 ...## ..$ PointMeasureCorr : num [1:3627] 0.42 0.42 0.3 0.27 0.31 0 0.14 -0.12 0.32 0 ...## ..$ Weight : num [1:3627] 1 1 1 1 1 1 1 1 1 1 ...## ..$ ObservMatch : num [1:3627] 75 80.6 91.7 97.2 86.1 100 94.4 50 97.2 100 ...## ..$ ExpectMatch : num [1:3627] 68.3 72 91.7 97.2 86.1 100 94.4 65 97.2 100 ...## ..$ PointMeasureExpected: num [1:3627] 0.35 0.33 0.2 0.12 0.25 0 0.17 0.38 0.17 0 ...## ..$ RMSR : num [1:3627] 0.42 0.39 0.26 0.16 0.31 0 0.23 0.51 0.14 0 ...## ..$ WMLE : num [1:3627] 1.25 1.68 3.13 4 2.59 ...## ..$ primary : Factor w/ 12 levels "0","10","20",..: 1 2 4 11 2 10 6 2 6 11 ...## ..$ secondary : Factor w/ 12 levels "0","10","20",..: 1 1 3 1 1 10 1 1 1 1 ...## $ : tibble[,26] [3,629 × 26] (S3: tbl_df/tbl/data.frame)## ..$ ssid : num [1:3629] 10129634 10926496 10616063 10139443 8887381 ...## ..$ grade : int [1:3629] 11 11 11 11 11 11 11 11 11 11 ...## ..$ year : int [1:3629] 18 18 18 18 18 18 18 18 18 18 ...## ..$ testeventid : num [1:3629] 147564 151249 146976 151229 147676 ...## ..$ asmtprmrydsbltycd : num [1:3629] 10 80 10 10 80 82 10 10 40 82 ...## ..$ asmtscndrydsbltycd : num [1:3629] 0 90 0 0 10 80 50 0 50 0 ...## ..$ Entry : num [1:3629] 161 344 321 167 101 131 141 357 14 367 ...## ..$ Theta : num [1:3629] 0.514 1.965 0.899 1.965 -4.724 ...## ..$ Status : num [1:3629] 1 1 1 1 -1 1 1 1 1 1 ...## ..$ Count : num [1:3629] 36 36 36 36 36 36 36 36 36 36 ...## ..$ RawScore : num [1:3629] 19 29 22 29 0 14 18 25 19 22 ...## ..$ SE : num [1:3629] 0.356 0.436 0.362 0.436 1.839 ...## ..$ Infit : num [1:3629] 0.97 1.03 0.99 1 1 1.01 0.99 0.85 0.96 1.06 ...## ..$ Infit_Z : num [1:3629] -0.18 0.26 -0.09 0.1 0 0.13 -0.06 -0.81 -0.39 0.5 ...## ..$ Outfit : num [1:3629] 0.97 1.03 1.01 0.87 1 0.93 1 0.77 0.97 0.99 ...## ..$ Outfit_Z : num [1:3629] -0.17 0.23 0.17 -0.46 0 -0.03 0.03 -0.87 -0.19 0.07 ...## ..$ Displacement : num [1:3629] 5e-04 5e-04 5e-04 5e-04 -7e-04 4e-04 5e-04 5e-04 5e-04 5e-04 ...## ..$ PointMeasureCorr : num [1:3629] 0.38 0.28 0.34 0.29 0 0.34 0.38 0.45 0.38 0.27 ...## ..$ Weight : num [1:3629] 1 1 1 1 1 1 1 1 1 1 ...## ..$ ObservMatch : num [1:3629] 66.7 80.6 66.7 80.6 100 61.1 69.4 75 63.9 61.1 ...## ..$ ExpectMatch : num [1:3629] 64.4 80.5 66 80.5 100 68.3 64.6 70.3 64.4 66 ...## ..$ PointMeasureExpected: num [1:3629] 0.35 0.25 0.33 0.25 0 0.35 0.35 0.3 0.35 0.33 ...## ..$ RMSR : num [1:3629] 0.46 0.39 0.45 0.39 0 0.46 0.46 0.4 0.46 0.48 ...## ..$ WMLE : num [1:3629] 0.512 1.915 0.888 1.915 -4.724 ...## ..$ primary : Factor w/ 12 levels "0","10","20",..: 2 10 2 2 10 11 2 2 4 11 ...## ..$ secondary : Factor w/ 12 levels "0","10","20",..: 1 12 1 1 2 10 6 1 6 1 ...## $ : tibble[,26] [3,627 × 26] (S3: tbl_df/tbl/data.frame)## ..$ ssid : num [1:3627] 18631185 18342736 10897771 7663935 6709613 ...## ..$ grade : int [1:3627] 11 11 11 11 11 11 11 11 11 11 ...## ..$ year : int [1:3627] 18 18 18 18 18 18 18 18 18 18 ...## ..$ testeventid : num [1:3627] 145641 147717 145196 149014 146545 ...## ..$ asmtprmrydsbltycd : num [1:3627] 10 90 10 80 0 10 10 80 82 82 ...## ..$ asmtscndrydsbltycd : num [1:3627] 0 0 0 10 0 82 0 70 0 0 ...## ..$ Entry : num [1:3627] 444 444 343 74 3 77 181 261 297 182 ...## ..$ Theta : num [1:3627] 2.24 3.48 2.72 -2.72 0.12 -1.39 -0.67 4.73 4.73 4.73 ...## ..$ Status : num [1:3627] 1 1 1 1 1 1 1 0 0 0 ...## ..$ Count : num [1:3627] 19 19 19 19 19 19 19 19 19 19 ...## ..$ RawScore : num [1:3627] 16 18 17 1 8 3 5 19 19 19 ...## ..$ SE : num [1:3627] 0.64 1.03 0.76 1.06 0.49 0.66 0.55 1.84 1.84 1.84 ...## ..$ Infit : num [1:3627] 1.07 1.02 1.1 1 0.98 0.73 1.08 1 1 1 ...## ..$ Infit_Z : num [1:3627] 0.29 0.31 0.53 0.29 -0.11 -0.54 0.37 0 0 0 ...## ..$ Outfit : num [1:3627] 1.17 0.94 1.22 0.76 0.95 0.62 1.09 1 1 1 ...## ..$ Outfit_Z : num [1:3627] 0.45 0.32 0.71 0 -0.22 -0.62 0.42 0 0 0 ...## ..$ Displacement : num [1:3627] 0 0 0 0 0 0 0 0 0 0 ...## ..$ PointMeasureCorr : num [1:3627] 0.01 0.08 0.05 0 0.32 0.68 0.2 0 0 0 ...## ..$ Weight : num [1:3627] 1 1 1 1 1 1 1 1 1 1 ...## ..$ ObservMatch : num [1:3627] 84.2 94.7 89.5 94.7 63.2 84.2 78.9 100 100 100 ...## ..$ ExpectMatch : num [1:3627] 84.2 94.7 89.5 94.7 64.2 85.1 76.3 100 100 100 ...## ..$ PointMeasureExpected: num [1:3627] 0.17 0.1 0.14 0.24 0.3 0.31 0.32 0 0 0 ...## ..$ RMSR : num [1:3627] 0.38 0.22 0.31 0.22 0.47 0.34 0.36 0 0 0 ...## ..$ WMLE : num [1:3627] 2.11 3.02 2.5 -2.27 0.14 -0.9 -0.6 4.73 4.73 4.73 ...## ..$ primary : Factor w/ 12 levels "0","10","20",..: 2 12 2 10 1 2 2 10 11 11 ...## ..$ secondary : Factor w/ 12 levels "0","10","20",..: 1 1 1 2 1 11 1 8 1 1 ...## $ : tibble[,26] [1,435 × 26] (S3: tbl_df/tbl/data.frame)## ..$ ssid : num [1:1435] 7617607 7642717 10341706 9811494 10469745 ...## ..$ grade : int [1:1435] 11 11 11 11 11 11 11 11 11 11 ...## ..$ year : int [1:1435] 18 18 18 18 18 18 18 18 18 18 ...## ..$ testeventid : num [1:1435] 148838 144146 149634 146456 144426 ...## ..$ asmtprmrydsbltycd : num [1:1435] 10 82 80 10 90 10 10 80 80 10 ...## ..$ asmtscndrydsbltycd : num [1:1435] 0 80 20 0 50 80 70 0 50 0 ...## ..$ Entry : num [1:1435] 28 43 274 118 291 91 58 130 297 337 ...## ..$ Theta : num [1:1435] 0.875 2.647 4.17 5.404 5.404 ...## ..$ Status : num [1:1435] 1 1 1 0 0 1 1 1 1 1 ...## ..$ Count : num [1:1435] 36 36 36 36 36 36 36 36 36 36 ...## ..$ RawScore : num [1:1435] 22 32 35 36 36 26 2 34 34 28 ...## ..$ SE : num [1:1435] 0.361 0.544 1.021 1.835 1.835 ...## ..$ Infit : num [1:1435] 1.3 0.94 1.05 1.04 0.99 0.9 0.97 0.99 0.98 0.9 ...## ..$ Infit_Z : num [1:1435] 2.35 0.02 0.37 0.35 0 -0.62 0.17 0.15 0.16 -0.52 ...## ..$ Outfit : num [1:1435] 1.22 0.76 1.5 1.2 0.48 0.85 0.69 1.57 0.67 0.8 ...## ..$ Outfit_Z : num [1:1435] 1.87 -0.27 0.91 0.6 -0.16 -0.87 -0.41 0.74 0.01 -0.76 ...## ..$ Displacement : num [1:1435] 0.0011 0.0016 0.0016 0.0027 0.0027 0.0013 0.0007 0.0016 0.0016 0.0014 ...## ..$ PointMeasureCorr : num [1:1435] 0.07 0.3 -0.07 0 0.29 0.46 0.28 0.05 0.22 0.45 ...## ..$ Weight : num [1:1435] 1 1 1 1 1 1 1 1 1 1 ...## ..$ ObservMatch : num [1:1435] 55.6 88.9 97.2 100 100 75 94.4 94.4 94.4 80.6 ...## ..$ ExpectMatch : num [1:1435] 66.1 88.9 97.2 100 100 73.2 94.4 94.4 94.4 77.9 ...## ..$ PointMeasureExpected: num [1:1435] 0.32 0.22 0.12 0 0 0.3 0.15 0.16 0.16 0.28 ...## ..$ RMSR : num [1:1435] 0.52 0.3 0.17 0 0 0.39 0.22 0.22 0.22 0.36 ...## ..$ WMLE : num [1:1435] 0.863 2.543 3.692 5.404 5.404 ...## ..$ primary : Factor w/ 12 levels "0","10","20",..: 2 11 10 2 12 2 2 10 10 2 ...## ..$ secondary : Factor w/ 12 levels "0","10","20",..: 1 10 3 1 6 10 8 1 6 1 ...## $ : tibble[,26] [3,627 × 26] (S3: tbl_df/tbl/data.frame)## ..$ ssid : num [1:3627] 7653093 7640498 7650957 9305807 18060455 ...## ..$ grade : int [1:3627] 11 11 11 11 11 11 11 11 11 11 ...## ..$ year : int [1:3627] 18 18 18 18 18 18 18 18 18 18 ...## ..$ testeventid : num [1:3627] 148893 148868 144397 148988 144865 ...## ..$ asmtprmrydsbltycd : num [1:3627] 10 10 90 0 82 10 80 0 10 82 ...## ..$ asmtscndrydsbltycd : num [1:3627] 0 0 80 0 0 0 50 0 0 10 ...## ..$ Entry : num [1:3627] 61 48 63 107 443 214 267 344 423 27 ...## ..$ Theta : num [1:3627] 1.67 2.17 -1.56 5.01 2.17 0.38 0.79 5.01 -0.48 2.78 ...## ..$ Status : num [1:3627] 1 1 1 0 1 1 1 0 1 1 ...## ..$ Count : num [1:3627] 13 13 13 13 13 13 13 13 13 13 ...## ..$ RawScore : num [1:3627] 9 10 2 13 10 6 7 13 4 11 ...## ..$ SE : num [1:3627] 0.69 0.74 0.83 1.87 0.74 0.64 0.65 1.87 0.68 0.84 ...## ..$ Infit : num [1:3627] 0.52 0.65 0.61 1 1.01 0.46 0.47 1 0.9 0.88 ...## ..$ Infit_Z : num [1:3627] -2.07 -1.02 -0.79 0 0.24 -2.3 -2.2 0 -0.16 -0.21 ...## ..$ Outfit : num [1:3627] 0.44 0.52 0.33 1 0.74 0.41 0.4 1 1.32 0.54 ...## ..$ Outfit_Z : num [1:3627] -1.65 -0.81 -0.47 0 -0.06 -1.88 -1.85 0 -0.14 -0.16 ...## ..$ Displacement : num [1:3627] 0 0 0 0 0 0 0 0 0 0 ...## ..$ PointMeasureCorr : num [1:3627] 0.86 0.73 0.62 0 0.51 0.87 0.87 0 0.56 0.56 ...## ..$ Weight : num [1:3627] 1 1 1 1 1 1 1 1 1 1 ...## ..$ ObservMatch : num [1:3627] 100 92.3 84.6 100 76.9 100 100 100 92.3 84.6 ...## ..$ ExpectMatch : num [1:3627] 75.5 79.7 84.6 100 79.7 72.9 73 100 74.5 84.6 ...## ..$ PointMeasureExpected: num [1:3627] 0.48 0.44 0.34 0 0.44 0.5 0.51 0 0.45 0.38 ...## ..$ RMSR : num [1:3627] 0.31 0.34 0.26 0 0.38 0.29 0.29 0 0.37 0.31 ...## ..$ WMLE : num [1:3627] 1.61 2.08 -1.38 5.01 2.08 0.38 0.78 5.01 -0.43 2.61 ...## ..$ primary : Factor w/ 12 levels "0","10","20",..: 2 2 12 1 11 2 10 1 2 11 ...## ..$ secondary : Factor w/ 12 levels "0","10","20",..: 1 1 10 1 1 1 6 1 1 2 ...It's a data frame!
We can add these summaries if we want
by_content %>% mutate(n = map_dbl(data, nrow))## # A tibble: 5 x 3## # Groups: content [5]## content data n## <chr> <list> <dbl>## 1 ELA <tibble[,26] [3,627 × 26]> 3627## 2 Math <tibble[,26] [3,629 × 26]> 3629## 3 Rdg <tibble[,26] [3,627 × 26]> 3627## 4 Science <tibble[,26] [1,435 × 26]> 1435## 5 Wri <tibble[,26] [3,627 × 26]> 3627map_*
Note on the previous example we used
map_dbland we got a vector in return.What would happen if we just used
map?
by_content %>% mutate(n = map(data, nrow))## # A tibble: 5 x 3## # Groups: content [5]## content data n ## <chr> <list> <list> ## 1 ELA <tibble[,26] [3,627 × 26]> <int [1]>## 2 Math <tibble[,26] [3,629 × 26]> <int [1]>## 3 Rdg <tibble[,26] [3,627 × 26]> <int [1]>## 4 Science <tibble[,26] [1,435 × 26]> <int [1]>## 5 Wri <tibble[,26] [3,627 × 26]> <int [1]>Let's fit a model!
by_content %>% mutate(m1 = map(data, ~lm(Theta ~ primary, data = .x)))## # A tibble: 5 x 3## # Groups: content [5]## content data m1 ## <chr> <list> <list>## 1 ELA <tibble[,26] [3,627 × 26]> <lm> ## 2 Math <tibble[,26] [3,629 × 26]> <lm> ## 3 Rdg <tibble[,26] [3,627 × 26]> <lm> ## 4 Science <tibble[,26] [1,435 × 26]> <lm> ## 5 Wri <tibble[,26] [3,627 × 26]> <lm>Extract the coefficients
by_content %>% mutate( m1 = map(data, ~lm(Theta ~ primary, data = .x)), coefs = map(m1, coef))## # A tibble: 5 x 4## # Groups: content [5]## content data m1 coefs ## <chr> <list> <list> <list> ## 1 ELA <tibble[,26] [3,627 × 26]> <lm> <dbl [11]>## 2 Math <tibble[,26] [3,629 × 26]> <lm> <dbl [12]>## 3 Rdg <tibble[,26] [3,627 × 26]> <lm> <dbl [11]>## 4 Science <tibble[,26] [1,435 × 26]> <lm> <dbl [12]>## 5 Wri <tibble[,26] [3,627 × 26]> <lm> <dbl [12]>by_content %>% mutate( m1 = map(data, ~lm(Theta ~ primary, data = .x)), coefs = map(m1, coef), no_disab = map_dbl(coefs, 1), tbi = no_disab + map_dbl(coefs, "primary74") ) %>% select(content, no_disab, tbi)## # A tibble: 5 x 3## # Groups: content [5]## content no_disab tbi## <chr> <dbl> <dbl>## 1 ELA 0.9322336 0.1674462## 2 Math -0.1587907 0.1910821## 3 Rdg 1.363101 1.629048 ## 4 Science 1.491319 2.790971 ## 5 Wri 1.571441 1.167429by_content %>% mutate( m1 = map(data, ~lm(Theta ~ primary, data = .x)), coefs = map(m1, coef), no_disab = map_dbl(coefs, 1), tbi = no_disab + map_dbl(coefs, "primary74") ) %>% select(content, no_disab, tbi)## # A tibble: 5 x 3## # Groups: content [5]## content no_disab tbi## <chr> <dbl> <dbl>## 1 ELA 0.9322336 0.1674462## 2 Math -0.1587907 0.1910821## 3 Rdg 1.363101 1.629048 ## 4 Science 1.491319 2.790971 ## 5 Wri 1.571441 1.167429Note - I wouldn't have neccesarily expected you to add no_disab to the TBI coefficient.
Model fits
mods <- by_content %>% mutate( m1 = map(data, ~lm(Theta ~ primary, data = .x)), m2 = map(data, ~lm(Theta ~ primary + secondary, data = .x)), m3 = map(data, ~lm(Theta ~ primary * secondary, data = .x)))mods## # A tibble: 5 x 5## # Groups: content [5]## content data m1 m2 m3 ## <chr> <list> <list> <list> <list>## 1 ELA <tibble[,26] [3,627 × 26]> <lm> <lm> <lm> ## 2 Math <tibble[,26] [3,629 × 26]> <lm> <lm> <lm> ## 3 Rdg <tibble[,26] [3,627 × 26]> <lm> <lm> <lm> ## 4 Science <tibble[,26] [1,435 × 26]> <lm> <lm> <lm> ## 5 Wri <tibble[,26] [3,627 × 26]> <lm> <lm> <lm>Brief foray into parallel iterations
The stats::anova function can compare the fit of two models
Pop Quiz
How would we extract just ELA model 1 and 2?
mods$m1[[1]]## ## Call:## lm(formula = Theta ~ primary, data = .x)## ## Coefficients:## (Intercept) primary10 primary20 primary40 primary50 primary60 ## 0.93223 0.38570 -0.03168 -1.84434 1.17372 0.77625 ## primary70 primary74 primary80 primary82 primary90 ## -1.83026 -0.76479 0.46765 0.33825 1.47936mods$m2[[1]]## ## Call:## lm(formula = Theta ~ primary + secondary, data = .x)## ## Coefficients:## (Intercept) primary10 primary20 primary40 primary50 primary60 ## 1.0043 0.4285 0.2807 -1.6282 1.1147 0.8676 ## primary70 primary74 primary80 primary82 primary90 secondary10 ## -1.4430 -0.1168 0.5283 0.3298 1.4101 -0.5685 ## secondary20 secondary40 secondary43 secondary50 secondary60 secondary70 ## -1.0558 -2.2707 -1.7943 0.2484 0.4831 -1.3336 ## secondary74 secondary80 secondary82 secondary90 ## -6.3695 -0.5142 -0.7259 1.6000Which fits better?
compare <- anova(mods$m1[[1]], mods$m2[[1]])compare## Analysis of Variance Table## ## Model 1: Theta ~ primary## Model 2: Theta ~ primary + secondary## Res.Df RSS Df Sum of Sq F Pr(>F) ## 1 3616 20905 ## 2 3605 20100 11 804.26 13.113 < 2.2e-16 ***## ---## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1map2
Works the same as
mapbut iterates over two vectors concurrentlyLet's compare model 1 and 2
mods %>% mutate(comp12 = map2(m1, m2, anova))## # A tibble: 5 x 6## # Groups: content [5]## content data m1 m2 m3 comp12 ## <chr> <list> <list> <list> <list> <list> ## 1 ELA <tibble[,26] [3,627 × 26]> <lm> <lm> <lm> <anova[,6] [2 × 6]>## 2 Math <tibble[,26] [3,629 × 26]> <lm> <lm> <lm> <anova[,6] [2 × 6]>## 3 Rdg <tibble[,26] [3,627 × 26]> <lm> <lm> <lm> <anova[,6] [2 × 6]>## 4 Science <tibble[,26] [1,435 × 26]> <lm> <lm> <lm> <anova[,6] [2 × 6]>## 5 Wri <tibble[,26] [3,627 × 26]> <lm> <lm> <lm> <anova[,6] [2 × 6]>map2
Works the same as
mapbut iterates over two vectors concurrentlyLet's compare model 1 and 2
mods %>% mutate(comp12 = map2(m1, m2, anova))## # A tibble: 5 x 6## # Groups: content [5]## content data m1 m2 m3 comp12 ## <chr> <list> <list> <list> <list> <list> ## 1 ELA <tibble[,26] [3,627 × 26]> <lm> <lm> <lm> <anova[,6] [2 × 6]>## 2 Math <tibble[,26] [3,629 × 26]> <lm> <lm> <lm> <anova[,6] [2 × 6]>## 3 Rdg <tibble[,26] [3,627 × 26]> <lm> <lm> <lm> <anova[,6] [2 × 6]>## 4 Science <tibble[,26] [1,435 × 26]> <lm> <lm> <lm> <anova[,6] [2 × 6]>## 5 Wri <tibble[,26] [3,627 × 26]> <lm> <lm> <lm> <anova[,6] [2 × 6]>Perhaps not terrifically helpful
Back to our anova object
- Can we pull out useful things?
str(compare)## Classes 'anova' and 'data.frame': 2 obs. of 6 variables:## $ Res.Df : num 3616 3605## $ RSS : num 20905 20100## $ Df : num NA 11## $ Sum of Sq: num NA 804## $ F : num NA 13.1## $ Pr(>F) : num NA 7.66e-25## - attr(*, "heading")= chr [1:2] "Analysis of Variance Table\n" "Model 1: Theta ~ primary\nModel 2: Theta ~ primary + secondary"Back to our anova object
- Can we pull out useful things?
str(compare)## Classes 'anova' and 'data.frame': 2 obs. of 6 variables:## $ Res.Df : num 3616 3605## $ RSS : num 20905 20100## $ Df : num NA 11## $ Sum of Sq: num NA 804## $ F : num NA 13.1## $ Pr(>F) : num NA 7.66e-25## - attr(*, "heading")= chr [1:2] "Analysis of Variance Table\n" "Model 1: Theta ~ primary\nModel 2: Theta ~ primary + secondary"Try pulling out the p value
All p-values
Note - this is probably the most compact syntax, but that doesn't mean it's the most clear
mods %>% mutate(comp12 = map2(m1, m2, anova), p12 = map_dbl(comp12, list("Pr(>F)", 2)))## # A tibble: 5 x 7## # Groups: content [5]## content data m1 m2 m3 comp12 p12## <chr> <list> <list> <list> <list> <list> <dbl>## 1 ELA <tibble[,26] [3,627 × 26]> <lm> <lm> <lm> <anova[,6] [2 × 6… 7.663566e-25## 2 Math <tibble[,26] [3,629 × 26]> <lm> <lm> <lm> <anova[,6] [2 × 6… 1.724262e-22## 3 Rdg <tibble[,26] [3,627 × 26]> <lm> <lm> <lm> <anova[,6] [2 × 6… 1.527172e-28## 4 Science <tibble[,26] [1,435 × 26]> <lm> <lm> <lm> <anova[,6] [2 × 6… 4.685885e-18## 5 Wri <tibble[,26] [3,627 × 26]> <lm> <lm> <lm> <anova[,6] [2 × 6… 5.785623e-11mods %>% mutate(comp12 = map2(m1, m2, anova), p12 = map_dbl(comp12, extract_p))## # A tibble: 5 x 7## # Groups: content [5]## content data m1 m2 m3 comp12 p12## <chr> <list> <list> <list> <list> <list> <dbl>## 1 ELA <tibble[,26] [3,627 × 26]> <lm> <lm> <lm> <anova[,6] [2 × 6… 7.663566e-25## 2 Math <tibble[,26] [3,629 × 26]> <lm> <lm> <lm> <anova[,6] [2 × 6… 1.724262e-22## 3 Rdg <tibble[,26] [3,627 × 26]> <lm> <lm> <lm> <anova[,6] [2 × 6… 1.527172e-28## 4 Science <tibble[,26] [1,435 × 26]> <lm> <lm> <lm> <anova[,6] [2 × 6… 4.685885e-18## 5 Wri <tibble[,26] [3,627 × 26]> <lm> <lm> <lm> <anova[,6] [2 × 6… 5.785623e-11Operations by row
The dplyr::rowwise() function fundamentally changes the way a tibble() behaves
df <- tibble(name = c("Me", "You"), x = 1:2, y = 3:4, z = 5:6)df %>% mutate(m = mean(c(x, y, z)))## # A tibble: 2 x 5## name x y z m## <chr> <int> <int> <int> <dbl>## 1 Me 1 3 5 3.5## 2 You 2 4 6 3.5df %>% rowwise() %>% mutate(m = mean(c(x, y, z)))## # A tibble: 2 x 5## # Rowwise: ## name x y z m## <chr> <int> <int> <int> <dbl>## 1 Me 1 3 5 3## 2 You 2 4 6 4List columns
If you apply rowwise operation with a list column, you don't have to loop
df <- tibble(x = list(1, 2:3, 4:6))df %>% mutate( l = map_int(x, length) )## # A tibble: 3 x 2## x l## <list> <int>## 1 <dbl [1]> 1## 2 <int [2]> 2## 3 <int [3]> 3df %>% rowwise() %>% mutate(l = length(x))## # A tibble: 3 x 2## # Rowwise: ## x l## <list> <int>## 1 <dbl [1]> 1## 2 <int [2]> 2## 3 <int [3]> 3Creating list columns
You can use the dplyr::nest_by() function to create a list column for each group, and convert it to a rowwise data frame.
d %>% nest_by(content)## # A tibble: 5 x 2## # Rowwise: content## content data## <chr> <list<tibble[,26]>>## 1 ELA [3,627 × 26]## 2 Math [3,629 × 26]## 3 Rdg [3,627 × 26]## 4 Science [1,435 × 26]## 5 Wri [3,627 × 26]Challenge
Given what we just learned, can you fit a model of the form Theta ~ primary to each content area (i.e., not using {purrr})?
02:00
list() (should suggest this in the error reporting if you don't)
d %>% nest_by(content) %>% mutate(m1 = list(lm(Theta ~ primary, data = data)))## # A tibble: 5 x 3## # Rowwise: content## content data m1 ## <chr> <list<tibble[,26]>> <list>## 1 ELA [3,627 × 26] <lm> ## 2 Math [3,629 × 26] <lm> ## 3 Rdg [3,627 × 26] <lm> ## 4 Science [1,435 × 26] <lm> ## 5 Wri [3,627 × 26] <lm>Challenge 2
Can you extend it further and extract the coefficients with coef? What about creating a new column that has the intercept values?
02:00
d %>% nest_by(content) %>% mutate(m1 = list(lm(Theta ~ primary, data = data)), coefs = list(coef(m1)))## # A tibble: 5 x 4## # Rowwise: content## content data m1 coefs ## <chr> <list<tibble[,26]>> <list> <list> ## 1 ELA [3,627 × 26] <lm> <dbl [11]>## 2 Math [3,629 × 26] <lm> <dbl [12]>## 3 Rdg [3,627 × 26] <lm> <dbl [11]>## 4 Science [1,435 × 26] <lm> <dbl [12]>## 5 Wri [3,627 × 26] <lm> <dbl [12]>Return atomic vectors
d %>% nest_by(content) %>% mutate(m1 = list(lm(Theta ~ primary, data = data)), intercept = coef(m1)[1])## # A tibble: 5 x 4## # Rowwise: content## content data m1 intercept## <chr> <list<tibble[,26]>> <list> <dbl>## 1 ELA [3,627 × 26] <lm> 0.9322336## 2 Math [3,629 × 26] <lm> -0.1587907## 3 Rdg [3,627 × 26] <lm> 1.363101 ## 4 Science [1,435 × 26] <lm> 1.491319 ## 5 Wri [3,627 × 26] <lm> 1.571441Fit all models
The below gets us the same results we got before
mods2 <- d %>% nest_by(content) %>% mutate( m1 = list(lm(Theta ~ primary, data = data)), m2 = list(lm(Theta ~ primary + secondary, data = data)), m3 = list(lm(Theta ~ primary * secondary, data = data)))mods2## # A tibble: 5 x 5## # Rowwise: content## content data m1 m2 m3 ## <chr> <list<tibble[,26]>> <list> <list> <list>## 1 ELA [3,627 × 26] <lm> <lm> <lm> ## 2 Math [3,629 × 26] <lm> <lm> <lm> ## 3 Rdg [3,627 × 26] <lm> <lm> <lm> ## 4 Science [1,435 × 26] <lm> <lm> <lm> ## 5 Wri [3,627 × 26] <lm> <lm> <lm>Look at all R2
It's a normal data frame!
mods %>% pivot_longer( m1:m3, names_to = "model", values_to = "output")## # A tibble: 15 x 4## # Groups: content [5]## content data model output## <chr> <list> <chr> <list>## 1 ELA <tibble[,26] [3,627 × 26]> m1 <lm> ## 2 ELA <tibble[,26] [3,627 × 26]> m2 <lm> ## 3 ELA <tibble[,26] [3,627 × 26]> m3 <lm> ## 4 Math <tibble[,26] [3,629 × 26]> m1 <lm> ## 5 Math <tibble[,26] [3,629 × 26]> m2 <lm> ## 6 Math <tibble[,26] [3,629 × 26]> m3 <lm> ## 7 Rdg <tibble[,26] [3,627 × 26]> m1 <lm> ## 8 Rdg <tibble[,26] [3,627 × 26]> m2 <lm> ## 9 Rdg <tibble[,26] [3,627 × 26]> m3 <lm> ## 10 Science <tibble[,26] [1,435 × 26]> m1 <lm> ## 11 Science <tibble[,26] [1,435 × 26]> m2 <lm> ## 12 Science <tibble[,26] [1,435 × 26]> m3 <lm> ## 13 Wri <tibble[,26] [3,627 × 26]> m1 <lm> ## 14 Wri <tibble[,26] [3,627 × 26]> m2 <lm> ## 15 Wri <tibble[,26] [3,627 × 26]> m3 <lm>Extract all R2
Note - might want to write a function here again
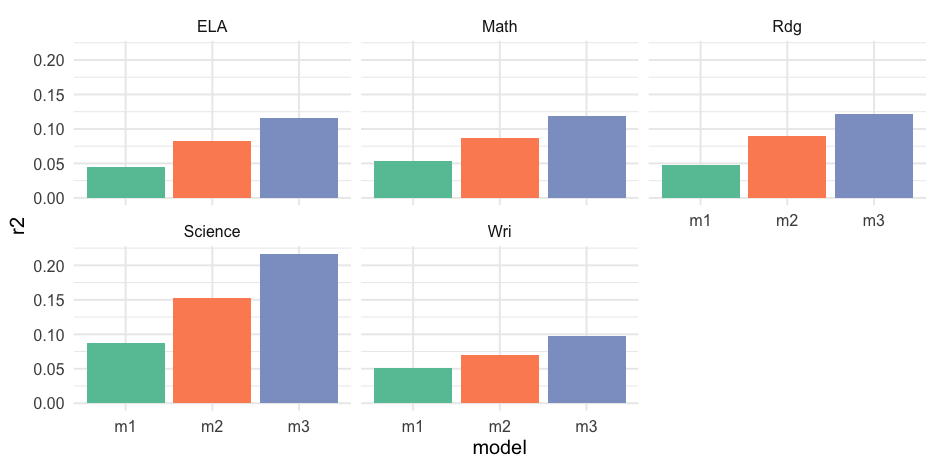
r2 <- mods %>% pivot_longer( m1:m3, names_to = "model", values_to = "output" ) %>% mutate(r2 = map_dbl(output, ~summary(.x)$r.squared))r2## # A tibble: 15 x 5## # Groups: content [5]## content data model output r2## <chr> <list> <chr> <list> <dbl>## 1 ELA <tibble[,26] [3,627 × 26]> m1 <lm> 0.04517421## 2 ELA <tibble[,26] [3,627 × 26]> m2 <lm> 0.08190917## 3 ELA <tibble[,26] [3,627 × 26]> m3 <lm> 0.1161187 ## 4 Math <tibble[,26] [3,629 × 26]> m1 <lm> 0.05326550## 5 Math <tibble[,26] [3,629 × 26]> m2 <lm> 0.08675264## 6 Math <tibble[,26] [3,629 × 26]> m3 <lm> 0.1185931 ## 7 Rdg <tibble[,26] [3,627 × 26]> m1 <lm> 0.04805713## 8 Rdg <tibble[,26] [3,627 × 26]> m2 <lm> 0.08926212## 9 Rdg <tibble[,26] [3,627 × 26]> m3 <lm> 0.1217497 ## 10 Science <tibble[,26] [1,435 × 26]> m1 <lm> 0.08683581## 11 Science <tibble[,26] [1,435 × 26]> m2 <lm> 0.1522437 ## 12 Science <tibble[,26] [1,435 × 26]> m3 <lm> 0.2170660 ## 13 Wri <tibble[,26] [3,627 × 26]> m1 <lm> 0.05171555## 14 Wri <tibble[,26] [3,627 × 26]> m2 <lm> 0.06977688## 15 Wri <tibble[,26] [3,627 × 26]> m3 <lm> 0.09820067Tidy
mods %>% pivot_longer( m1:m3, names_to = "model", values_to = "output" ) %>% mutate(tidied = map(output, broom::tidy))## # A tibble: 15 x 5## # Groups: content [5]## content data model output tidied ## <chr> <list> <chr> <list> <list> ## 1 ELA <tibble[,26] [3,627 × 26]> m1 <lm> <tibble[,5] [11 × 5]> ## 2 ELA <tibble[,26] [3,627 × 26]> m2 <lm> <tibble[,5] [22 × 5]> ## 3 ELA <tibble[,26] [3,627 × 26]> m3 <lm> <tibble[,5] [132 × 5]>## 4 Math <tibble[,26] [3,629 × 26]> m1 <lm> <tibble[,5] [12 × 5]> ## 5 Math <tibble[,26] [3,629 × 26]> m2 <lm> <tibble[,5] [23 × 5]> ## 6 Math <tibble[,26] [3,629 × 26]> m3 <lm> <tibble[,5] [144 × 5]>## 7 Rdg <tibble[,26] [3,627 × 26]> m1 <lm> <tibble[,5] [11 × 5]> ## 8 Rdg <tibble[,26] [3,627 × 26]> m2 <lm> <tibble[,5] [22 × 5]> ## 9 Rdg <tibble[,26] [3,627 × 26]> m3 <lm> <tibble[,5] [132 × 5]>## 10 Science <tibble[,26] [1,435 × 26]> m1 <lm> <tibble[,5] [12 × 5]> ## 11 Science <tibble[,26] [1,435 × 26]> m2 <lm> <tibble[,5] [22 × 5]> ## 12 Science <tibble[,26] [1,435 × 26]> m3 <lm> <tibble[,5] [132 × 5]>## 13 Wri <tibble[,26] [3,627 × 26]> m1 <lm> <tibble[,5] [12 × 5]> ## 14 Wri <tibble[,26] [3,627 × 26]> m2 <lm> <tibble[,5] [22 × 5]> ## 15 Wri <tibble[,26] [3,627 × 26]> m3 <lm> <tibble[,5] [132 × 5]>Equivalently
mods %>% pivot_longer( m1:m3, names_to = "model", values_to = "output" ) %>% rowwise() %>% mutate(tidied = list(broom::tidy(output)))## # A tibble: 15 x 5## # Rowwise: content## content data model output tidied ## <chr> <list> <chr> <list> <list> ## 1 ELA <tibble[,26] [3,627 × 26]> m1 <lm> <tibble[,5] [11 × 5]> ## 2 ELA <tibble[,26] [3,627 × 26]> m2 <lm> <tibble[,5] [22 × 5]> ## 3 ELA <tibble[,26] [3,627 × 26]> m3 <lm> <tibble[,5] [132 × 5]>## 4 Math <tibble[,26] [3,629 × 26]> m1 <lm> <tibble[,5] [12 × 5]> ## 5 Math <tibble[,26] [3,629 × 26]> m2 <lm> <tibble[,5] [23 × 5]> ## 6 Math <tibble[,26] [3,629 × 26]> m3 <lm> <tibble[,5] [144 × 5]>## 7 Rdg <tibble[,26] [3,627 × 26]> m1 <lm> <tibble[,5] [11 × 5]> ## 8 Rdg <tibble[,26] [3,627 × 26]> m2 <lm> <tibble[,5] [22 × 5]> ## 9 Rdg <tibble[,26] [3,627 × 26]> m3 <lm> <tibble[,5] [132 × 5]>## 10 Science <tibble[,26] [1,435 × 26]> m1 <lm> <tibble[,5] [12 × 5]> ## 11 Science <tibble[,26] [1,435 × 26]> m2 <lm> <tibble[,5] [22 × 5]> ## 12 Science <tibble[,26] [1,435 × 26]> m3 <lm> <tibble[,5] [132 × 5]>## 13 Wri <tibble[,26] [3,627 × 26]> m1 <lm> <tibble[,5] [12 × 5]> ## 14 Wri <tibble[,26] [3,627 × 26]> m2 <lm> <tibble[,5] [22 × 5]> ## 15 Wri <tibble[,26] [3,627 × 26]> m3 <lm> <tibble[,5] [132 × 5]>Select and unnest
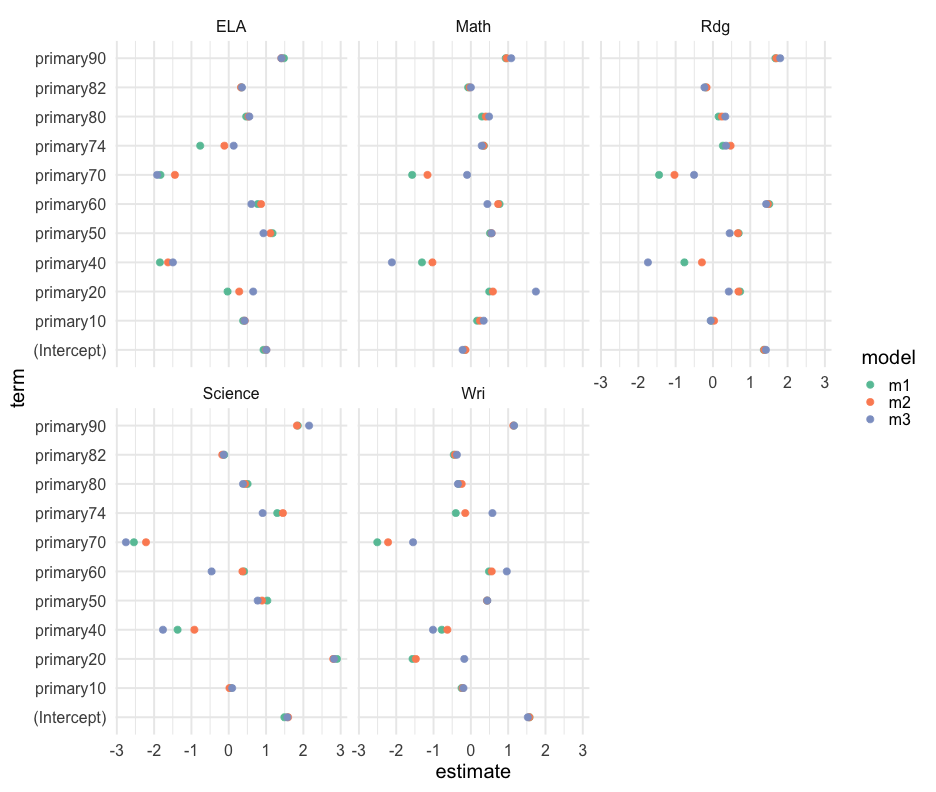
tidied <- mods %>% gather(model, output, m1:m3) %>% mutate(tidied = map(output, broom::tidy)) %>% select(content, model, tidied) %>% unnest(tidied)tidied## # A tibble: 841 x 7## # Groups: content [5]## content model term estimate std.error statistic p.value## <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>## 1 ELA m1 (Intercept) 0.9322336 0.2150561 4.334839 1.498396e-5## 2 ELA m1 primary10 0.3856986 0.2242965 1.719593 8.559207e-2## 3 ELA m1 primary20 -0.03167527 0.7266436 -0.04359120 9.652327e-1## 4 ELA m1 primary40 -1.844343 0.5559031 -3.317741 9.164595e-4## 5 ELA m1 primary50 1.173722 0.2890447 4.060694 4.996391e-5## 6 ELA m1 primary60 0.7762539 0.3866313 2.007737 4.474555e-2## 7 ELA m1 primary70 -1.830257 0.3086128 -5.930595 3.301860e-9## 8 ELA m1 primary74 -0.7647874 0.5182670 -1.475663 1.401215e-1## 9 ELA m1 primary80 0.4676481 0.2428640 1.925556 5.423822e-2## 10 ELA m1 primary82 0.3382547 0.2267600 1.491686 1.358687e-1## # … with 831 more rowsBy grade
by_grade_content <- d %>% group_by(content, grade) %>% nest()by_grade_content## # A tibble: 31 x 3## # Groups: grade, content [31]## grade content data ## <int> <chr> <list> ## 1 11 ELA <tibble[,25] [453 × 25]>## 2 11 Math <tibble[,25] [460 × 25]>## 3 11 Rdg <tibble[,25] [453 × 25]>## 4 11 Science <tibble[,25] [438 × 25]>## 5 11 Wri <tibble[,25] [453 × 25]>## 6 3 ELA <tibble[,25] [540 × 25]>## 7 3 Math <tibble[,25] [536 × 25]>## 8 3 Rdg <tibble[,25] [540 × 25]>## 9 3 Wri <tibble[,25] [540 × 25]>## 10 4 ELA <tibble[,25] [585 × 25]>## # … with 21 more rowsFit models
mods_grade <- by_grade_content %>% mutate( m1 = map(data, ~lm(Theta ~ primary, data = .x)), m2 = map(data, ~lm(Theta ~ primary + secondary, data = .x)), m3 = map(data, ~lm(Theta ~ primary * secondary, data = .x)))mods_grade## # A tibble: 31 x 6## # Groups: grade, content [31]## grade content data m1 m2 m3 ## <int> <chr> <list> <list> <list> <list>## 1 11 ELA <tibble[,25] [453 × 25]> <lm> <lm> <lm> ## 2 11 Math <tibble[,25] [460 × 25]> <lm> <lm> <lm> ## 3 11 Rdg <tibble[,25] [453 × 25]> <lm> <lm> <lm> ## 4 11 Science <tibble[,25] [438 × 25]> <lm> <lm> <lm> ## 5 11 Wri <tibble[,25] [453 × 25]> <lm> <lm> <lm> ## 6 3 ELA <tibble[,25] [540 × 25]> <lm> <lm> <lm> ## 7 3 Math <tibble[,25] [536 × 25]> <lm> <lm> <lm> ## 8 3 Rdg <tibble[,25] [540 × 25]> <lm> <lm> <lm> ## 9 3 Wri <tibble[,25] [540 × 25]> <lm> <lm> <lm> ## 10 4 ELA <tibble[,25] [585 × 25]> <lm> <lm> <lm> ## # … with 21 more rowsLook at R2
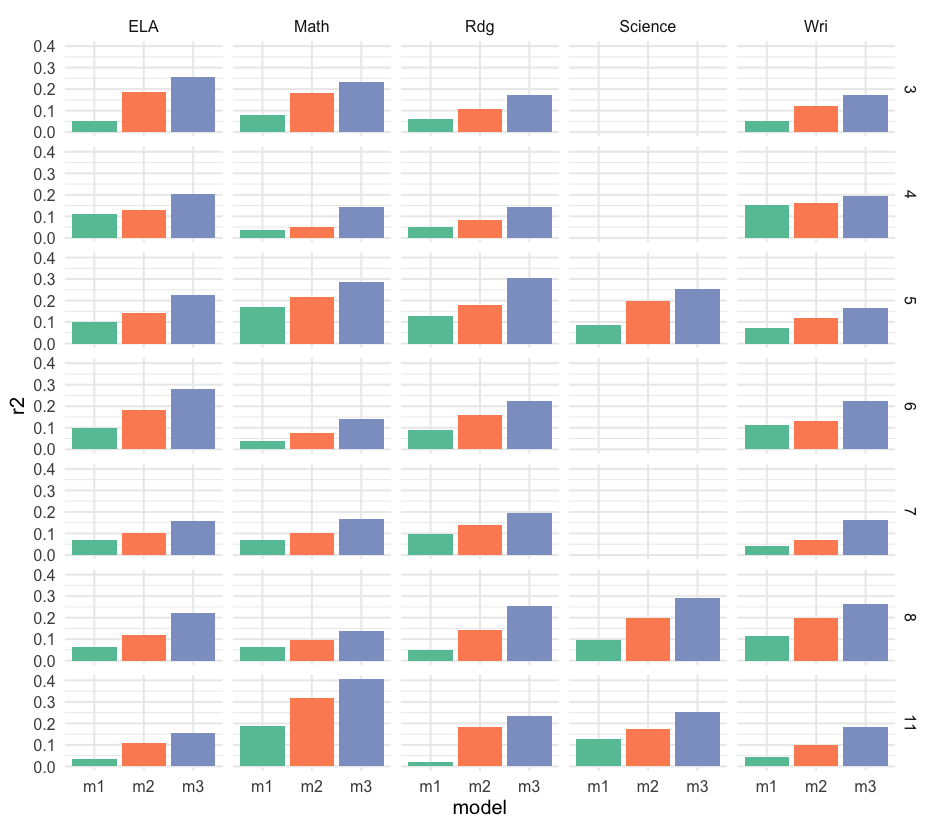
mods_grade %>% pivot_longer( m1:m3, names_to = "model", values_to = "output" ) %>% mutate(r2 = map_dbl(output, ~summary(.x)$r.squared))## # A tibble: 93 x 6## # Groups: grade, content [31]## grade content data model output r2## <int> <chr> <list> <chr> <list> <dbl>## 1 11 ELA <tibble[,25] [453 × 25]> m1 <lm> 0.03353818## 2 11 ELA <tibble[,25] [453 × 25]> m2 <lm> 0.1084394 ## 3 11 ELA <tibble[,25] [453 × 25]> m3 <lm> 0.1536891 ## 4 11 Math <tibble[,25] [460 × 25]> m1 <lm> 0.1886003 ## 5 11 Math <tibble[,25] [460 × 25]> m2 <lm> 0.3161226 ## 6 11 Math <tibble[,25] [460 × 25]> m3 <lm> 0.4046634 ## 7 11 Rdg <tibble[,25] [453 × 25]> m1 <lm> 0.02066316## 8 11 Rdg <tibble[,25] [453 × 25]> m2 <lm> 0.1820512 ## 9 11 Rdg <tibble[,25] [453 × 25]> m3 <lm> 0.2337721 ## 10 11 Science <tibble[,25] [438 × 25]> m1 <lm> 0.1259080 ## # … with 83 more rowsSummary
List columns are really powerful and really flexible
Also help you stay organized
You can approach the problem either with {purrr} or
dplyr::rowwise().Important: If you use
rowwise(), remember toungroup()when you want it to go back to being a normal data frameI'm asking you to learn both - the row-wise approach might be a bit easier but is a little less general (only works with data frames)