Functions: Part 3
Daniel Anderson
Week 7, Class 2
What does this mean operationally?
Your function should do ONE thing (and do it well)
Careful when naming functions - be as clear as possible
Embed useful error messages and warnings
- Particularly if you're working on a package or set of functions or others are using your functions
Refactor your code to be more clear after initial drafts (it's okay to be messy on a first draft)
Example 1
- Anything we can do to clean this up?
both_na <- function(x, y) { if(length(x) != length(y)) { lx <- length(x) ly <- length(y) v_lngths <- paste0("x = ", lx, ", y = ", ly) if(lx %% ly == 0 | ly %% lx == 0) { warning("Vectors were recycled (", v_lngths, ")") } else { stop("Vectors are of different lengths and are not recyclable:", v_lngths) } } sum(is.na(x) & is.na(y))}Revision
both_na <- function(x, y) { if(!recyclable(x, y)) { stop("Vectors are of different lengths and are not recyclable: ", "(x = ", length(x), ", y = ", length(y), ")") } if(length(x) == length(y)) { return(sum(is.na(x) & is.na(y))) } if(recyclable(x, y)) { warning("Vectors were recycled (", "x = ", length(x), ", y = ", length(y), ")") return(sum(is.na(x) & is.na(y))) }}Test it
both_na(a, b)## [1] 2both_na(a, c(b, b))## Warning in both_na(a, c(b, b)): Vectors were recycled (x = 7, y = 14)## [1] 4both_na(c(a, b), c(b, b, b))## Error in both_na(c(a, b), c(b, b, b)): Vectors are of different lengths and are not recyclable: (x = 14, y = 21)both_na(c(a, a), b)## Warning in both_na(c(a, a), b): Vectors were recycled (x = 14, y = 7)## [1] 4Anything else?
Make errors/warnings a function
check_lengths <- function(x, y) { if(!length(x) == length(y)) { if(recyclable(x, y)) { warning("Vectors were recycled (", "x = ", length(x), ", y = ", length(y), ")") } else { stop("Vectors are of different lengths and are not recyclable: ", "(x = ", length(x), ", y = ", length(y), ")") } }}Test it
both_na(a, b)## [1] 2both_na(a, c(b, b))## Warning in check_lengths(x, y): Vectors were recycled (x = 7, y = 14)## [1] 4both_na(c(a, b), c(b, b, b))## Error in check_lengths(x, y): Vectors are of different lengths and are not recyclable: (x = 14, y = 21)both_na(c(a, a), b)## Warning in check_lengths(x, y): Vectors were recycled (x = 14, y = 7)## [1] 4What is NSE
Implementation of different scoping rules
In dplyr and many others, arguments are evaluated inside the specified data frames, rather than the current or global environment.
How?
(a) Capture an expression (quote it) (b) Use the expression within the correct context (evaluate it)
So, x is evaluated as, e.g., df$x rather than globalenv()$x.
Example
Using the percentile_df function we created previously
- Here
base::substitute
percentile_df <- function(x) { sorted <- sort(x) d <- data.frame(sorted, percentile = ecdf(sorted)(sorted)) names(d)[1] <- paste0(substitute(x), collapse = "_") d}percentile_df(rnorm(100, 5, 0.2)) %>% head()## rnorm_100_5_0.2 percentile## 1 4.384718 0.01## 2 4.494705 0.02## 3 4.500262 0.03## 4 4.562786 0.04## 5 4.564927 0.05## 6 4.568238 0.06Actually getting this thing to work
extract_var <- function(df, var) { subset(eval(substitute(df)), select = var)}extract_var(mtcars, "mpg")## mpg## Mazda RX4 21.0## Mazda RX4 Wag 21.0## Datsun 710 22.8## Hornet 4 Drive 21.4## Hornet Sportabout 18.7## Valiant 18.1## Duster 360 14.3## Merc 240D 24.4## Merc 230 22.8## Merc 280 19.2## Merc 280C 17.8## Merc 450SE 16.4## Merc 450SL 17.3## Merc 450SLC 15.2## Cadillac Fleetwood 10.4## Lincoln Continental 10.4## Chrysler Imperial 14.7## Fiat 128 32.4## Honda Civic 30.4## Toyota Corolla 33.9## Toyota Corona 21.5## Dodge Challenger 15.5## AMC Javelin 15.2## Camaro Z28 13.3## Pontiac Firebird 19.2## Fiat X1-9 27.3## Porsche 914-2 26.0## Lotus Europa 30.4## Ford Pantera L 15.8## Ferrari Dino 19.7## Maserati Bora 15.0## Volvo 142E 21.4Better
Use NSE for both arguments
extract_var <- function(df, var) { eval(substitute(var), envir = df)}extract_var(mtcars, mpg)## [1] 21.0 21.0 22.8 21.4 18.7 18.1 14.3 24.4 22.8 19.2 17.8 16.4 17.3 15.2 10.4 10.4 14.7 32.4 30.4 33.9 21.5 15.5 15.2 13.3 19.2 27.3 26.0 30.4 15.8 19.7 15.0 21.4Better
Use NSE for both arguments
extract_var <- function(df, var) { eval(substitute(var), envir = df)}extract_var(mtcars, mpg)## [1] 21.0 21.0 22.8 21.4 18.7 18.1 14.3 24.4 22.8 19.2 17.8 16.4 17.3 15.2 10.4 10.4 14.7 32.4 30.4 33.9 21.5 15.5 15.2 13.3 19.2 27.3 26.0 30.4 15.8 19.7 15.0 21.4The above is equivalent to
df$varbut where bothdfandvarcan be swapped programmatically.The
varargument is being substituted in for whatever the user supplies, and is being evaluated within thedfenvironment
Maybe more simply
extract_var <- function(df, var) { df[ ,as.character(substitute(var))]}extract_var(mtcars, mpg)## [1] 21.0 21.0 22.8 21.4 18.7 18.1 14.3 24.4 22.8 19.2 17.8 16.4 17.3 15.2 10.4 10.4 14.7 32.4 30.4 33.9 21.5 15.5 15.2 13.3 19.2 27.3 26.0 30.4 15.8 19.7 15.0 21.4Why
as.character? Otherwise it is a symbol, which won't work.Note we could add
drop = FALSEto this if we wanted to maintain the data frame structure
Taking this even further
extract_vars <- function(df, ...) { vars <- substitute(alist(...)) df[ ,as.character(vars)[-1]]}head(extract_vars(mtcars, mpg, cyl, disp))## mpg cyl disp## Mazda RX4 21.0 6 160## Mazda RX4 Wag 21.0 6 160## Datsun 710 22.8 4 108## Hornet 4 Drive 21.4 6 258## Hornet Sportabout 18.7 8 360## Valiant 18.1 6 225We've now basically replicated
dplyr::selectNotice the use of
[-1], because callingas.characteronvarsalways returnsalistas the first element in the vector
Why is NSE used so frequently in the tidyverse?
mpg %>% select(manufacturer, model, hwy)## # A tibble: 234 x 3## manufacturer model hwy## <chr> <chr> <int>## 1 audi a4 29## 2 audi a4 29## 3 audi a4 31## 4 audi a4 30## 5 audi a4 26## 6 audi a4 26## 7 audi a4 27## 8 audi a4 quattro 26## 9 audi a4 quattro 25## 10 audi a4 quattro 28## # … with 224 more rowsmtcars %>% group_by(cyl, gear) %>% summarize(mean = mean(mpg, na.rm = TRUE)) %>% pivot_wider(names_from = cyl, values_from = mean)## # A tibble: 3 x 4## gear `4` `6` `8`## <dbl> <dbl> <dbl> <dbl>## 1 3 21.5 19.75 15.05## 2 4 26.925 19.75 NA ## 3 5 28.2 19.7 15.4- Try generalizing the above code into a function
04:00
But it doesn't...
group_means(mtcars, mpg, cyl, gear)## Error: Must group by variables found in `.data`.## * Column `group_1` is not found.## * Column `group_2` is not found.group_means(diamonds, price, cut, clarity)## Error: Must group by variables found in `.data`.## * Column `group_1` is not found.## * Column `group_2` is not found.But it doesn't...
group_means(mtcars, mpg, cyl, gear)## Error: Must group by variables found in `.data`.## * Column `group_1` is not found.## * Column `group_2` is not found.group_means(diamonds, price, cut, clarity)## Error: Must group by variables found in `.data`.## * Column `group_1` is not found.## * Column `group_2` is not found.Why?
- It's looking for an object called
group_1that doesn't exist inside the function or in the global workspace!
The {rlang} version
group_means <- function(data, outcome, group_1, group_2) { out <- enquo(outcome) # Quote the inputs g1 <- enquo(group_1) g2 <- enquo(group_2) data %>% group_by(!!g1, !!g2) %>% # !! to evaluate (bang-bang) summarize(mean = mean(!!out, na.rm = TRUE)) %>% pivot_wider(names_from = !!g1, values_from = mean)}group_means(mtcars, mpg, cyl, gear)## # A tibble: 3 x 4## gear `4` `6` `8`## <dbl> <dbl> <dbl> <dbl>## 1 3 21.5 19.75 15.05## 2 4 26.925 19.75 NA ## 3 5 28.2 19.7 15.4group_means(diamonds, price, cut, clarity)## # A tibble: 8 x 6## clarity Fair Good `Very Good` Premium Ideal## <ord> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 I1 3703.533 3596.635 4078.226 3947.332 4335.726## 2 SI2 5173.916 4580.261 4988.688 5545.937 4755.953## 3 SI1 4208.279 3689.533 3932.391 4455.269 3752.118## 4 VS2 4174.724 4262.236 4215.760 4550.331 3284.550## 5 VS1 4165.141 3801.446 3805.353 4485.462 3489.744## 6 VVS2 3349.768 3079.108 3037.765 3795.123 3250.290## 7 VVS1 3871.353 2254.774 2459.441 2831.206 2468.129## 8 IF 1912.333 4098.324 4396.216 3856.143 2272.913Alternative: Pass the dots!
- Note, I've made the function a bit simpler here by removing the spread
group_means2 <- function(data, outcome, ...) { out <- enquo(outcome) # Still have to quote the outcome data %>% group_by(...) %>% summarize(mean = mean(!!out, na.rm = TRUE)) }group_means2(mtcars, mpg, cyl, gear)## # A tibble: 8 x 3## # Groups: cyl [3]## cyl gear mean## <dbl> <dbl> <dbl>## 1 4 3 21.5 ## 2 4 4 26.925## 3 4 5 28.2 ## 4 6 3 19.75 ## 5 6 4 19.75 ## 6 6 5 19.7 ## 7 8 3 15.05 ## 8 8 5 15.4group_means2(diamonds, price, cut, clarity)## # A tibble: 40 x 3## # Groups: cut [5]## cut clarity mean## <ord> <ord> <dbl>## 1 Fair I1 3703.533## 2 Fair SI2 5173.916## 3 Fair SI1 4208.279## 4 Fair VS2 4174.724## 5 Fair VS1 4165.141## 6 Fair VVS2 3349.768## 7 Fair VVS1 3871.353## 8 Fair IF 1912.333## 9 Good I1 3596.635## 10 Good SI2 4580.261## # … with 30 more rowsAdded benefit
I can now also pass as many columns as I want, and it will still work!
group_means2(diamonds, price, cut, clarity, color)## # A tibble: 276 x 4## # Groups: cut, clarity [40]## cut clarity color mean## <ord> <ord> <ord> <dbl>## 1 Fair I1 D 7383 ## 2 Fair I1 E 2095.222## 3 Fair I1 F 2543.514## 4 Fair I1 G 3187.472## 5 Fair I1 H 4212.962## 6 Fair I1 I 3501 ## 7 Fair I1 J 5795.043## 8 Fair SI2 D 4355.143## 9 Fair SI2 E 4172.385## 10 Fair SI2 F 4520.112## # … with 266 more rowsPipe-centric
- Because the
dataargument comes first, this function works just like any other in the tidyverse, e.g.,
diamonds %>% filter(color == "E") %>% select(carat, cut, clarity) %>% group_means3(carat, cut, clarity)## # A tibble: 8 x 6## clarity Fair Good `Very Good` Premium Ideal## <ord> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 I1 0.9688889 1.330870 1.069545 1.043 1.037778 ## 2 SI2 1.015641 0.8825743 0.9304045 0.9576686 0.8744136## 3 SI1 0.8670769 0.7238592 0.7230831 0.7262866 0.6706266## 4 VS2 0.6902381 0.739375 0.6644135 0.6189348 0.5211356## 5 VS1 0.6328571 0.6806742 0.6097952 0.6431507 0.5035919## 6 VVS2 0.6007692 0.5601923 0.4267114 0.5115702 0.4839053## 7 VVS1 0.64 0.4181395 0.4000588 0.4622857 0.4265075## 8 IF NA 0.3733333 0.5793023 0.5762963 0.4577215Challenge
Write a function that summarizes any numeric columns by returning the mean, standard deviation, min, and max values.
For bonus points, embed a meaningful error message if the columns supplied are not numeric.
Example
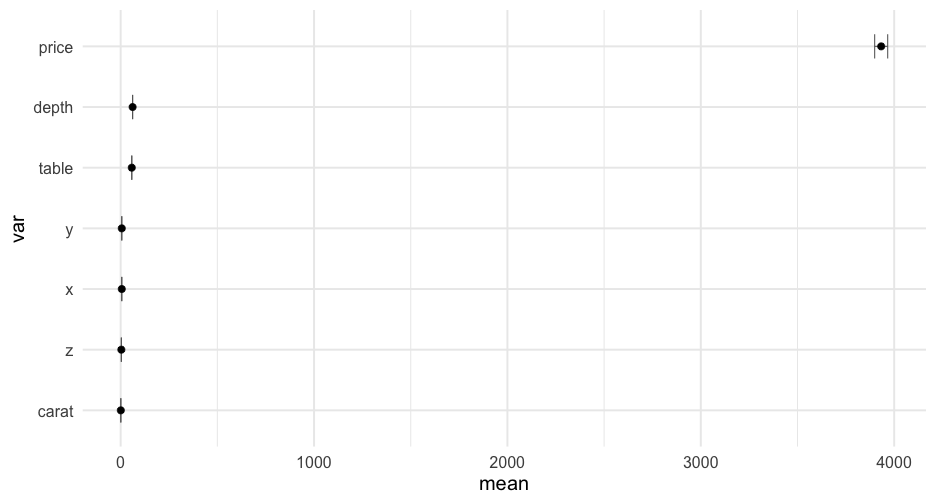
summarize_cols(diamonds, depth, table, price)## # A tibble: 3 x 5## var mean sd min max## <chr> <dbl> <dbl> <dbl> <dbl>## 1 depth 61.74940 1.432621 43 79## 2 price 3932.800 3989.440 326 18823## 3 table 57.45718 2.234491 43 9507:00
Pass the dots!
summarize_cols <- function(data, ...) { data %>% select(...) %>% pivot_longer(everything(), names_to = "var", values_to = "val") %>% group_by(var) %>% summarize(mean = mean(val, na.rm = TRUE), sd = sd(val, na.rm = TRUE), min = min(val, na.rm = TRUE), max = max(val, na.rm = TRUE))}Pipe-centric again
Just putting data as the first argument leads to a lot of benefits
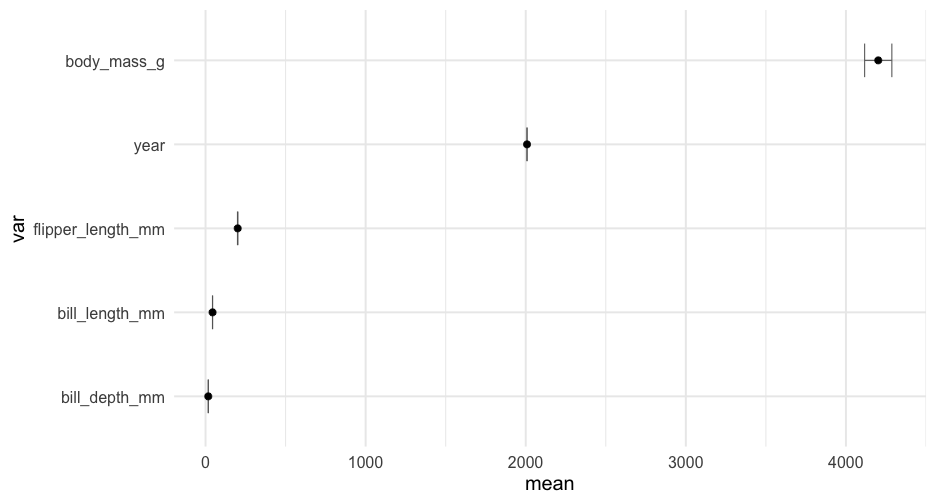
library(palmerpenguins)penguins %>% select_if(is.numeric) %>% summarize_cols(everything())## # A tibble: 5 x 5## var mean sd min max## <chr> <dbl> <dbl> <dbl> <dbl>## 1 bill_depth_mm 17.15117 1.974793 13.1 21.5## 2 bill_length_mm 43.92193 5.459584 32.1 59.6## 3 body_mass_g 4201.754 801.9545 2700 6300 ## 4 flipper_length_mm 200.9152 14.06171 172 231 ## 5 year 2008.029 0.8183559 2007 2009Maybe this?
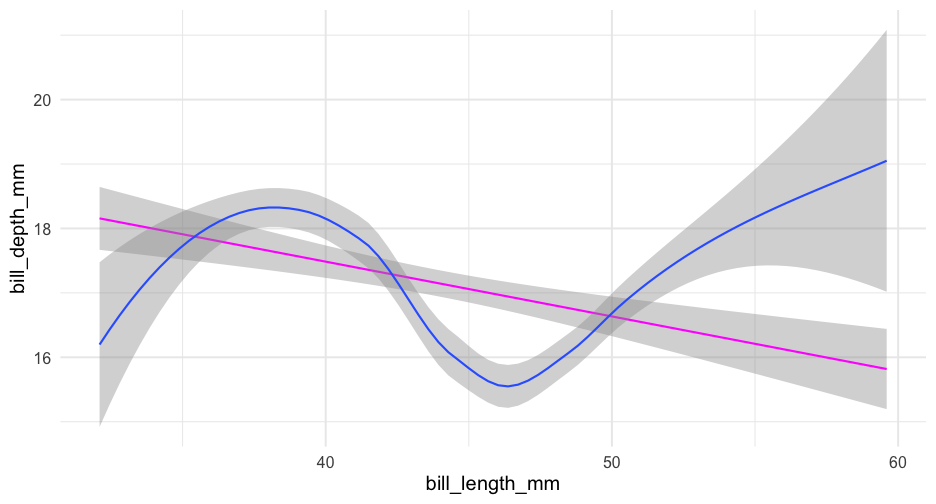
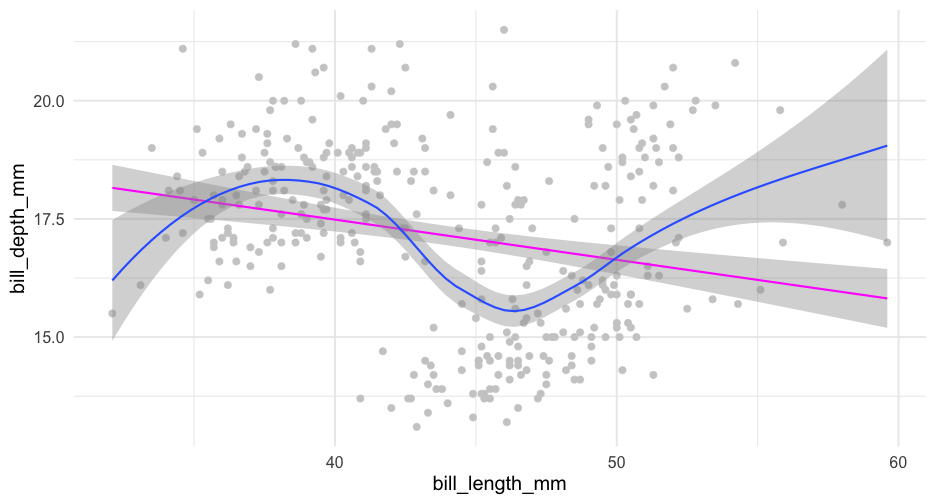
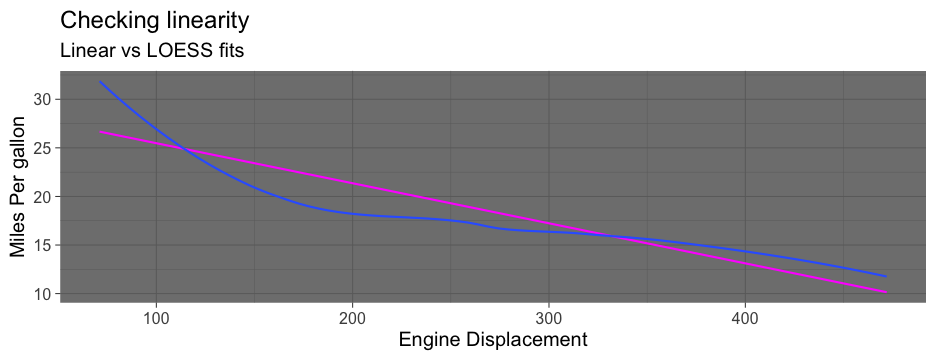
check_linear <- function(data, x, y, se = TRUE, points = FALSE) { p <- ggplot(data, aes(x, y)) if(points) { p <- p + geom_point(color = "gray80") } if(se) { p <- p + geom_smooth(method = "lm") + geom_smooth() } else { p <- p + geom_smooth(method = "lm", se = FALSE) + geom_smooth(se = FALSE) } p}Use tidyeval
Note - there are other approaches too, but they are soft deprecated
check_linear <- function(data, x, y, points = FALSE, ...) { p <- ggplot(data, aes({{x}}, {{y}})) if (points) { p <- p + geom_point(color = "gray80") } p + geom_smooth(method = "lm", color = "magenta", ...) + geom_smooth(...)}Full means/SEs
penguins %>% select_if(is.numeric) %>% pivot_longer(everything(), names_to = "var", values_to = "val") %>% group_by(var) %>% summarize(mean = mean(val, na.rm = TRUE), se = se(val))## # A tibble: 5 x 3## var mean se## <chr> <dbl> <dbl>## 1 bill_depth_mm 17.15117 0.1067846 ## 2 bill_length_mm 43.92193 0.2952205 ## 3 body_mass_g 4201.754 43.36473 ## 4 flipper_length_mm 200.9152 0.7603704 ## 5 year 2008.029 0.04412279Translate to a function
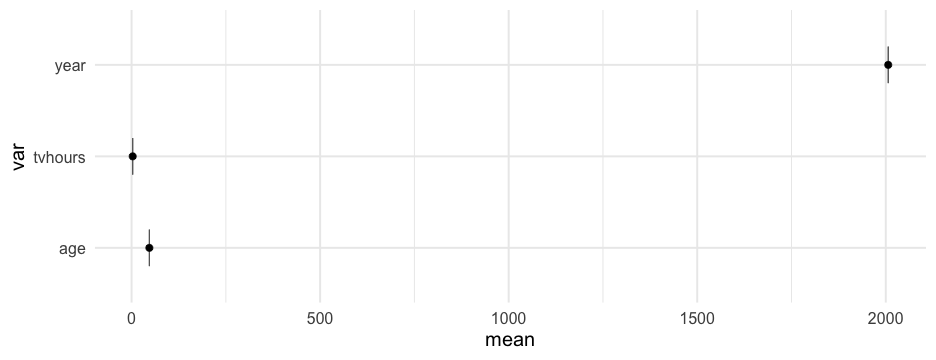
estimate_means <- function(df) { df %>% select_if(is.numeric) %>% pivot_longer(everything(), names_to = "var", values_to = "val") %>% group_by(var) %>% summarize(mean = mean(val, na.rm = TRUE), se = se(val)) }estimate_means(forcats::gss_cat)## # A tibble: 3 x 3## var mean se## <chr> <dbl> <dbl>## 1 age 47.18008 0.1181556 ## 2 tvhours 2.980771 0.02429812## 3 year 2006.502 0.03037436Create plot function
plot_means <- function(df) { means <- estimate_means(df) %>% mutate(var = reorder(factor(var), mean)) ggplot(means, aes(var, mean)) + geom_errorbar(aes(ymin = mean + qnorm(0.025)*se, ymax = mean + qnorm(0.975)*se), width = 0.4, color = "gray40") + geom_point() + coord_flip()}Create plot function
plot_means <- function(df) { means <- estimate_means(df) %>% mutate(var = reorder(factor(var), mean)) ggplot(means, aes(var, mean)) + geom_errorbar(aes(ymin = mean + qnorm(0.025)*se, ymax = mean + qnorm(0.975)*se), width = 0.4, color = "gray40") + geom_point() + coord_flip()}Notice we've successfully avoided tideval entirely in both examples!